Kim, Youn, Chang, Kang, and Jeon: Identification of Differentially-Methylated Genes and Pathways in Patients with Delayed Cerebral Ischemia Following Subarachnoid Hemorrhage
Abstract
Objective
We reported the differentially methylated genes in patients with subarachnoid hemorrhage (SAH) using bioinformatics analyses to explore the biological characteristics of the development of delayed cerebral ischemia (DCI).
Methods
DNA methylation profiles obtained from 40 SAH patients from an epigenome-wide association study were analyzed. Functional enrichment analysis, protein-protein interaction (PPI) network, and module analyses were carried out.
Results
A total of 13 patients (32.5%) experienced DCI during the follow-up. In total, we categorized the genes into the two groups of hypermethylation (n=910) and hypomethylation (n=870). The hypermethylated genes referred to biological processes of organic cyclic compound biosynthesis, nucleobase-containing compound biosynthesis, heterocycle biosynthesis, aromatic compound biosynthesis and cellular nitrogen compound biosynthesis. The hypomethylated genes referred to biological processes of carbohydrate metabolism, the regulation of cell size, and the detection of a stimulus, and molecular functions of amylase activity, and hydrolase activity. Based on PPI network and module analysis, three hypermethylation modules were mainly associated with antigen-processing, Golgi-to-ER retrograde transport, and G alpha (i) signaling events, and two hypomethylation modules were associated with post-translational protein phosphorylation and the regulation of natural killer cell chemotaxis. VHL, KIF3A, KIFAP3, RACGAP1, and OPRM1 were identified as hub genes for hypermethylation, and ALB and IL5 as hub genes for hypomethylation.
Conclusion
This study provided novel insights into DCI pathogenesis following SAH. Differently methylated hub genes can be useful biomarkers for the accurate DCI diagnosis.
Key Words: Subarachnoid hemorrhage · Intracranial aneurysm · DNA methylation · Computational biology · Epigenetics · Delayed cerebral ischemia.
INTRODUCTION
Delayed cerebral ischemia (DCI) refers to the clinical syndrome of newly developed focal neurologic deficits following an aneurysmal subarachnoid hemorrhage (SAH). It usually starts from 3 days and persists until 2 weeks after ictus [ 3, 31]. The improper management of DCI is the most important cause of poor neurologic outcome following SAH, although successful treatment for a ruptured aneurysm itself is done by surgical clipping or endovascular coil embolization [ 4, 31]. DCI was thought to be solely associated with cerebral vasospasm in the past. However, other causes such as early brain injury, cortical spreading depolarization or microcirculatory dysfunction were also correlated with DCI [ 6]. Accordingly, DCI is regarded as a much more complex disease than just narrowing of the large cerebral artery [ 6]. Genetic studies performed to investigate DCI pathogenesis have focused on the frequency of candidate gene variants or differential DNA methylation of CpG sites. Starke et al. [ 35] reported that DCI risk was well-correlated with the T allele of endothelial nitric oxide synthase compared to the C allele (odds ratio [OR], 10.9 for TT and 3.3 for one T allele). Kim et al. [ 11] showed higher methylation of the inositol 1-,4-,5-trisphosphate receptor (ITPR3) with lower ITPR3 mRNA expression in DCI patients. A recent epigenome-wide association study (EWAS) revealed two DCI-related CpG hypermethylation sites, the insulin receptor ( INSR) gene and the cadherin-related family member 5 ( CDHR5) [ 13]. However, more detailed bioinformatics analyses were not performed based on the degree of methylation. Bioinformatics analyses have been increasingly reported to identify novel molecular pathways in disease pathogenesis [ 33]. Sang et al. [ 33] revealed various epigenetic hub genes, such as PTGS2, PIK3CD, CXCL1, ESR1, and MMP2, which were associated with hepatocellular carcinoma. Herein, we evaluated the biological characteristics of SAH patients with DCI based on differentially methylated genes to provide novel insights into DCI pathogenesis.
MATERIALS AND METHODS
Bioinformatics analyses obtained based on a prospective cohort study were approved by the Institutional Review Board (No. 2017-9, 2018-6, and 2019-6) of Hallym University Chuncheon Sacred Heart Hospital. Informed consent was obtained from the patients or their relatives.
Study characteristics
In this study, we aimed to perform functional analyses for DCI development using various bioinformatics tools. DCI referred to the following conditions : 1) newly occurring neurologic abnormalities, such as motor weakness, sensory changes, dysphasia, or decreased levels of consciousness; 2) any changes greater than 2 points in the Glasgow coma scale or National Institutes of Health stroke scale; and 3) the presence of severe angiographic vasospasm, defined as arterial narrowing over 50% [ 11, 12, 35]. Clinical information (e.g., gender, age, hypertension, diabetes mellitus, hyperlipidemia, and smoking history) and radiologic findings were reviewed.
Processing of DNA methylation data
We analyzed further the EWAS data sets of SAH patients after reporting methylated genes related to DCI at the EWAS level ( Supplementary Methods) [ 13]. The data involved an epigenome-wide methylation profile of CpG sites in 40 patients with aneurysmal SAH using the Infinium MethylationEPIC (EPIC) BeadChip (Illumina, San Diego, CA, USA) [ 13]. Raw quality probes in the raw data were filtered by the minfi package (version 1.24.0). First, the samples with detection p-values of >0.05 were discarded. Then, the remaining probes were normalized using the preprocess Funnorm algorithm implemented in the minfi package. This method eliminates undesirable variation by regressing out variability with the control probes present in our methylation microarray. Finally, the differentially methylated CpG sites (DMCpGs) were identified with a q-value cutoff of 0.01. Among the identified DMCpGs, CpG sites showing a difference in beta (β) value greater than 0.2 were used for further analysis. Finally, we selected genes with DMCpGs located in their promoter regions (±2 kb of transcription start site) and used them to infer the potential functions of the DMCpGs in terms of gene regulation.
Bioinformatics analyses
Functional enrichment analysis was carried out using the web-based gene enrichment analysis and visualization tool ( http://cbl-gorilla.cs.technion.ac.il). Then, we constructed a protein-protein interaction (PPI) network using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING). The standard cutoff was regarded as an interaction score of 0.4 [ 33]. Cluster-ONE application was used for module analysis. A module was defined as the number of connected interactions over 10. The hub genes were defined as genes with high connectivity ranked in the top 10% of the candidate modules [ 23].
RESULTS
Identification of differentially methylated genes
The clinical characteristics of the SAH patients extracted from the previously reported studies are presented in Supplementary Table 1 [ 13]. All patients enrolled in EWAS underwent surgical clipping or endovascular coiling within 12 hours after ictus. Among them, 13 patients (32.5%) experienced DCI during the follow-up period. The number of hypermethylated and hypomethylated genes were 910 and 870, respectively. Hierarchical clustering of CpG sites according to differences in DNA methylation is described in the Supplementary Fig. 1.
Functional enrichment analysis
Among the retrieved gene ontology (GO) biological processes (BP) involved in hypermethylation, those associated with the organic cyclic compound biosynthetic process, the nucleobase-containing compound biosynthetic process, the heterocyclic compound biosynthetic process, the aromatic compound biosynthetic process, and the cellular nitrogen compound biosynthetic process were the most significant ( Table 1 and Fig. 1). The retrieved GO terms of BP in hypomethylation were the carbohydrate metabolic process, the regulation of cell size, and the detection of a stimulus. The GO molecular function (MF) terms were amylase activity and hydrolyzing O-glycosyl compounds ( Table 1 and Fig. 1).
PPI network and module analysis
In total, 367 nodes (genes) and 756 edges (interactions) in hypermethylation and 360 nodes and 644 edges in hypomethylation were established. The PPI networks for hypermethylation and hypomethylation are shown in the Supplementary Fig. 2 and 3, respectively. Three modules (PPI-based protein clusters) in the hypermethylated genes ( Fig. 2) and two modules in the hypomethylated genes ( Fig. 3) were constructed with statistical significance. Enrichment analyses showed that the hypermethylation modules were mainly associated with antigen-processing (ubiquitination and proteasome degradation), golgi-to-ER retrograde transport, and G alpha (i) signaling events. Two hypermethylation modules were closely correlated with post-translational protein phosphorylation and the regulation of natural killer cell chemotaxis. The five hypermethylated hub genes included von Hippel-Lindau ( VHL), kinesin family member ( KIF) 3A, kinesin associated protein 3 ( KIFAP3), Rac GTPase activating protein 1 ( RACGAP1), and opioid receptor mu 1 ( OPRM1). The two hypomethylated hub genes were albumin ( ALB) and interleukin 5 ( IL5) ( Table 2).
DISCUSSION
The development of DCI showed a dynamic nature that was involved with multiple molecular changes. Most studies have focused on DCI-associated proteomic biomarkers or single nucleotide polymorphisms in candidate genes [ 19]. Lad et al. [ 19] reported that tumor necrosis factor-α, interleukin-1, soluble tumor necrosis factor receptor I, and neurofilament proteins were reliable markers for the diagnosis of DCI. A genomewide association study revealed that DCI was closely associated with SNP rs999662, which encompasses solute carrier family 12 member 3 [ 14]. More specifically, the thymine-thymine of rs999662 was associated with a higher risk of DCI compared to guanine-thymine ( p=8.69×10 -6). Epigenetic mechanisms include DNA methylation and histone post-translational modifications [ 34]. Epigenetic analysis in stroke research has usually been performed for atherosclerosis and ischemic stroke. Zaina et al. [ 37] compared DNA methylation changes in atherosclerosis between atherosclerotic human and donor-matched healthy aortas. Differentially methylated genes were closely related to endothelial and vascular smooth muscle cell function. Endres et al. [ 5] reported that DNA methyltransferase ( DNMTs) gene deletion mice were resistant to mild ischemic brain injury, indicating that DNMT activity was associated with the aggravation of brain inury after middle cerebral artery occlusion (MCAO). Regarding SAH, epigenetic mechanisms have not been well studied. Kim et al. [ 11] investigated the association between methylation in the distal intergenic region ( IGR) of ITPR3 and gene expression. DCI patients had increased levels of methylation intensity compared to non-DCI patients (DCI, 0.941 [0.857-0.984] vs. nonDCI, 0.670 [0.543-0.761]) with increased DNMT1 and decreased ten-eleven translocation enzyme ( TET) 1 expression. They hypothesized that hypermethylation of the distal IGR by increased DNMT1 and decreased TET1 repressed ITPR3 gene expression following initial SAH ictus [ 11]. Recently, an association between the hypermethylation of CpG sites in cg00441765 ( INSR gene) and cg11464053 ( CDHR5 gene) with decreased mRNA expression and DCI development was reported [ 13]. Nikkola et al. [ 26] observed 164 differentially expressed genes and 3,493 differently methylated CpG sites in SAH patients who underwent remote ischemic conditioning. The gene enrichment and PPI analyses in their study revealed 204 CpG sites overlapped with 103 genes and relevant pathways of the cell cycle and inflammatory responses. However, remote ischemic conditioning refers to the protection of subsequent injuries though repetitive ischemic exposure in the peripheral tissues. Accordingly, bioinformatics in SAH patients with remote ischemic conditioning cannot accurately reflect the pathogenesis of DCI. In our study, bioinformatics analyses disclosed several novel differentially methylated hub genes, which were closely associated with DCI. The mutational inactivation of VHL or hypermethylation of a normally unmethylated CpG island in the 5’ region was observed in clear cell renal cell carcinoma [ 9, 25]. VHL is an essential component of the ubiquitin-proteasome degradation pathway, which is responsible for the suppression of hypoxia-inducible transcription factor (HIF) in normoxic condition [ 20]. Lei et al. [ 20] demonstrated that chronic activation of the HIF 1 pathway in mice with cardiac myocyte-specific deletion of VHL can lead to cardiac degeneration. KIF3A was reported to be related to protein and nucleotide bindings and microtubule motor activity [ 16]. Kovacic et al. [ 16] reported that the T allele of KIF3A rs7737031 significantly increased the risk of asthma. In addition, KIF3A was downregulated in patients with asthma, suggesting its role in a diminished lung’s ability to remove inhaled particles. Guinot et al. [ 7] demonstrated that the combined deletion of KIF3A and VHL as well as TRP53 caused cystic and neoplastic change in the kidney. DNA methylation at the CpG site of the OPRM1 promoter with decreased gene expression was associated with perioperative pain in patients who underwent spinal fusion [ 2]. Opioids can alter the tight junction protein expression in the blood-brain barrier, aggravating stroke pathogenesis [ 29]. Opioid dependency was an independent risk factor for stroke development [ 8]. However, endogenous opioids showed cardioprotection during ischemic conditions by opening the mitochondrial K + ATP channels [ 29, 30]. Accordingly, further studies focusing on the role of OPRM1 hypermethylation in the development of DCI are necessary. Two hub genes of hypomethylation, IL5 and ALB, were observed in our study. IL5 has shown a protective effect in atherosclerosis [ 15]. Zhao et al. [ 38] reported that the inhibition of macrophage-specific overexpression of IL5 was associated with the progression of atherosclerotic lesions. Regarding acute stroke patients, higher levels of IL5 were related to lesser stroke severity and favorable outcomes [ 21]. Serum ALB was also correlated with post-stroke outcomes [ 1]. Babu et al. [ 1] reported that lower ALB levels increased the risk of poor neurologic outcomes (OR, 1.972; 95% confidence interval [CI], 1.103-4.001). In addition, significantly decreased ALB levels were observed in recurrence or death in patients with ischemic stroke. Suarez et al. [ 36] showed a neuroprotective effect of 25% human ALB with dosage of 1.25 g/kg/day compared to 0.625 g/kg/day (OR, 3.0513; 95% CI, 0.6586-14.1367) in patients with SAH. The expression of ALB mRNA was increased in riboflavin-deficient cells by a loss of lysine-specific demethylase 1 activity and the subsequent accumulation of lysine-4-demethylated histone H3 marks in the promoter of ALB in human hepatocarcinoma cells [ 22]. Undermethylation of the 5’ end of the ALB gene was required for ALB production in rat hepatoma cells [ 27]. Nevertheless, the effect of a hypomethylated ALB genes on DCI remains unclear and requires additional study for the precise mechanisms involved in DCI pathogenesis. BP enrichment analysis of the hypermethylated genes included the cellular nitrogen compound biosynthetic process. Glutamine and glutamate are used as nitrogen donors for the synthesis of nitrogenous compounds in cells [ 17]. Samuelsson et al. [ 32] reported that increased interstitial glutamine was associated with improved cerebral perfusion pressure and decreased intracranial pressure, suggesting augmented astrocyte energy metabolism by increases in glutamate uptake and glutamine synthesis. Increased glutamate oxidation and metabolism of glutamine was also demonstrated in neuronal and glial recovery following focal cerebral ischemia [ 28]. Regarding the hypomethylated genes in SAH patients with DCI, BP, and MF enrichment analyses results included the carbohydrate metabolic process and alpha-amylase activity. Acute stroke patients tend to experience disorders of carbohydrate metabolism, in particular, hemorrhage stroke [ 24]. Hyperglycemia has been associated with stroke severity, neurological decline, and mortality compared to euglycemia [ 18]. The level of saliva alpha-amylase activity in stroke patients was increased compared to healthy controls [ 10]. In summary, bioinformatics analyses allow a better understanding of biological and metabolic processes in DCI pathogenesis. Nevertheless, the differentiation of pathological pathways between the causes and results of DCI development remains unsolved. Therefore, additional conjoint analysis using DNA methylation and gene expression is necessary to reveal more accurate biological functions involved in DCI pathogenesis [ 33]. Our study had some limitations. First, the small number of enrolled patients is a concern, although this was the bioinformatics analysis of epigenetic changes in SAH patients with DCI. Second, gene expression data were not involved in this analysis. Alternative splicing was observed in both hyper- and hypomethylated genes. Thus, additional epigenetic studies joining gene expression data are required in a large SAH cohort.
CONCLUSION
We found BP and hub genes that were closely associated with DCI in patients with SAH using various bioinformatics tools. Hypermethylated hub genes such as VHL, KIF3A, KIFAP3, RACGAP1, and OPRM1, and hypomethylated hub genes, including ALB and IL5, can be feasibly used to accurately diagnose DCI that requires timely treatment. Further studies focusing on possible gene regulatory control from the perspective of epigenetics are required.
Acknowledgements
This research was supported by the National Research Foundation of Korea funded by the Ministry of Education (2020R1l1A3070726) and Hallym University Research Fund.
Supplementary materials
Supplementary Table 1.
Clinical characteristics of the SAH patients enrolled in these bioinformatics analyses
Supplementary Fig. 1.
Hierarchical clustering of differences in DNA methylation according to delayed cerebral ischemia. The identification numbers of CpG site are presented in detail in the attached excel file.
Supplemental Fig. 2.
Protein-protein interaction network of hypermethylated genes between subarachnoid hemorrhage patients with and without delayed cerebral ischemia.
Supplemental Fig. 3.
Protein-protein interaction network of hypomethylated genes between subarachnoid hemorrhage patients with and without delayed cerebral ischemia.
Fig. 1.
Functional enrichment analyses of hypermethylated (A) and hypomethylated genes (B) between subarachnoid hemorrhage patients with and without delayed cerebral ischemia. The blue-colored boxes indicate statistical significance. 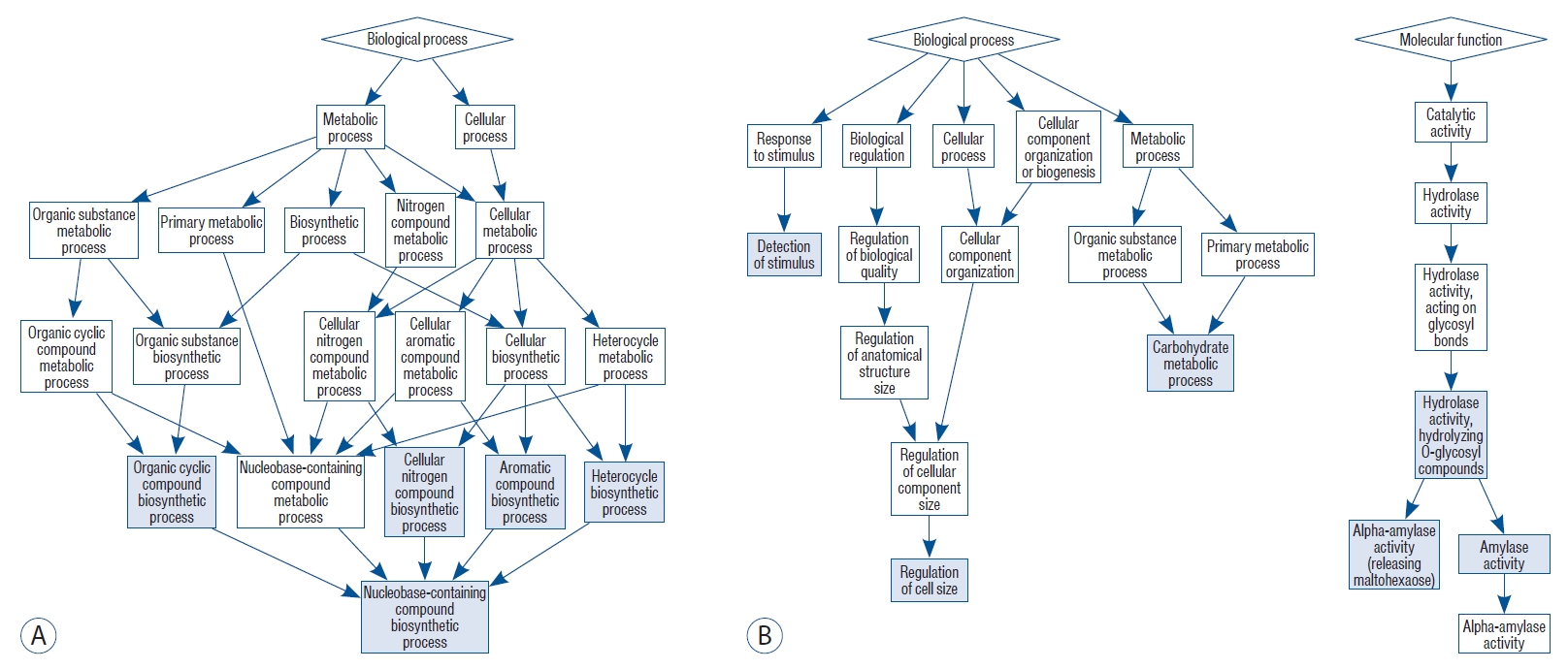
Fig. 2.
Module cluster analyses of hypermethylated genes and related gene enrichment analysis between delayed cerebral ischemia (DCI) and non-DCI patients following subarachnoid hemorrhage. 
Fig. 3.
Module cluster analyses of hypomethylated genes and related gene enrichment analysis between delayed cerebral ischemia (DCI) and non-DCI patients following subarachnoid hemorrhage. 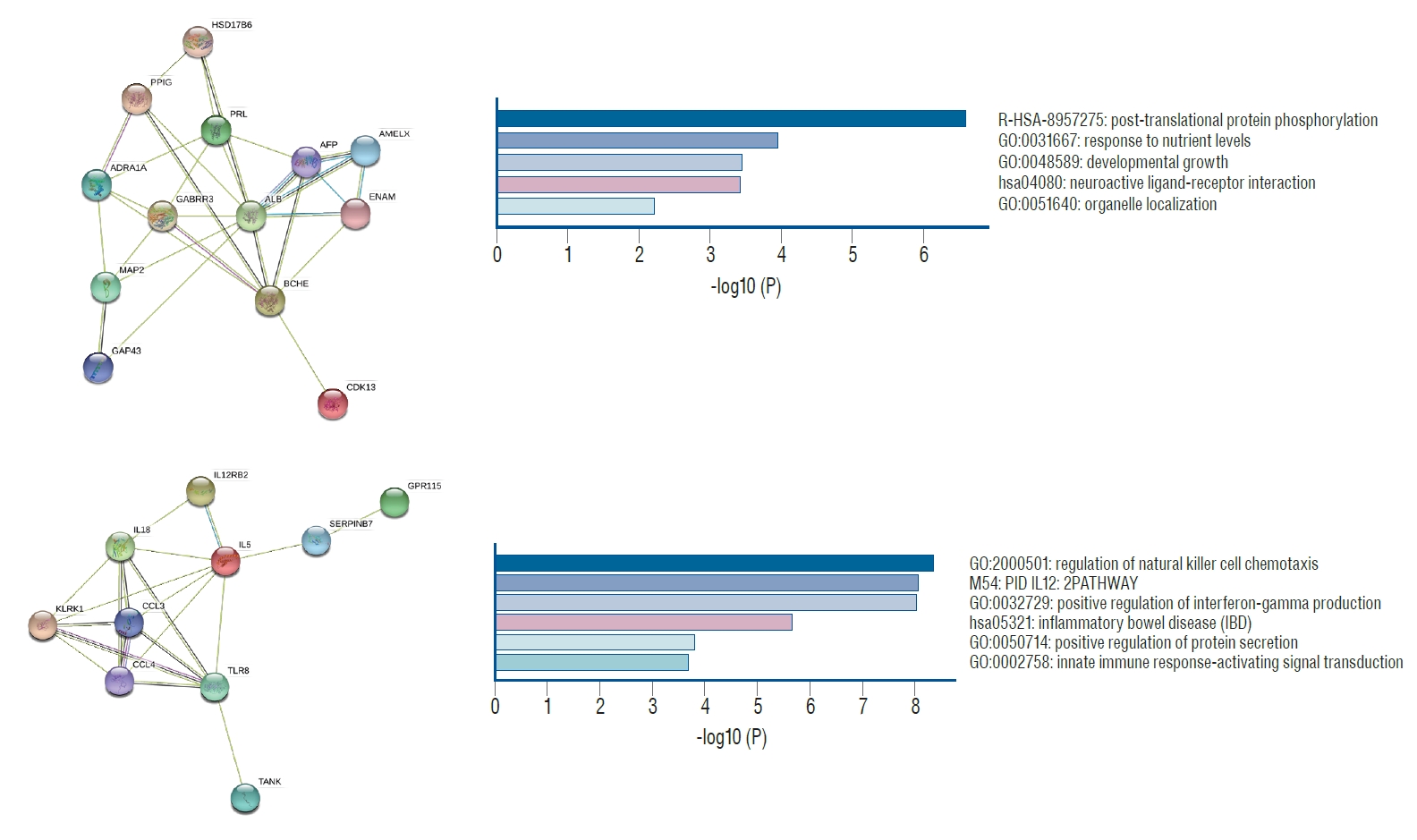
Table 1.
Functional enrichment analyses of differentially methylated genes between SAH patients with and without DCI
|
Category |
ID |
Description |
p-value |
Gene |
|
Hypermethylation |
|
|
|
|
|
BP |
GO:1901362 |
Organic cyclic compound biosynthetic process |
7.73E-05 |
DLAT, ADCY10, ERCC6, ACAT1, FUBP1, AK5, EGR2, LSM10, PTEN, HSD3B2, RRAGC, SMG7, THRAP3, PPT1
|
|
GO:0034654 |
Nucleobase-containing compound biosynthetic process |
1.49E-04 |
DLAT, ADCY10, ACAT1, ERCC6, FUBP1, AK5, EGR2, LSM10, PTEN, RRAGC, SMG7, THRAP3, PPT1
|
|
GO:0018130 |
Heterocycle biosynthetic process |
1.49E-04 |
DLAT, ADCY10, ACAT1, ERCC6, FUBP1, AK5, EGR2, LSM10, PTEN, RRAGC, SMG7, THRAP3, PPT1
|
|
GO:0019438 |
Aromatic compound biosynthetic process |
1.49E-04 |
DLAT, ADCY10, ACAT1, ERCC6, FUBP1, AK5, EGR2, LSM10, PTEN, RRAGC, SMG7, THRAP3, PPT1
|
|
GO:0044271 |
Cellular nitrogen compound biosynthetic process |
2.17E-04 |
RPS24, DLAT, ADCY10, ERCC6, ACAT1, FUBP1, AK5, EGR2, LSM10, PTEN, RRAGC, ASAH2, SMG7, THRAP3, PPT1
|
|
Hypomethylation |
|
|
|
|
|
BP |
GO:0005975 |
Carbohydrate metabolic process |
5.01E-05 |
AMY2B, AMY2A
|
|
GO:0008361 |
Regulation of cell size |
2.41E-04 |
CLCN6, MTOR
|
|
GO:0051606 |
Detection of stimulus |
6.71E-04 |
RYR2, OR6A2, OR51G2, OR2T12, OR5AR1, OR5A1, OR5D18, OR8U1, OR2AK2, OR14A16, PDC, OR6N1, TAS1R3, OR14I1
|
|
GO:0004556 |
Amylase activity |
5.15E-05 |
AMY2B, AMY2A
|
|
MT |
GO:0004553 |
Hydrolase activity, hydrolyzing O-glycosyl compounds |
8.96E-04 |
AMY2B, AMY2A
|
Table 2.
Gene module analysis of protein-protein interaction network based on the degree of methyaltion between DCI and non-DCI patients following SAH
|
Methylation |
Module |
Size |
p-value |
Gene |
|
Hypermethylation |
1 |
14 |
9.75E-05 |
VHL, ASB15, ASB18, KLHL5, MGRN1, PARK2, RNF19A, LONRF1, UBE2G1, SOCS5, CCDC182, SHISA9, SLITRK6, USP33
|
|
2 |
12 |
8.60E-05 |
KIF3A, KIFAP3, RACGAP1, SEC22B, COPA, DCTN4, HLA-DOB, HLA-DQB1, KIF21A, NBAS, TAP1, AGPAT1
|
|
3 |
10 |
0.025917 |
OPRM1, ADORA3, GNB4, C5, TAS2R16, NPL, GPM6A, LTBP1, NCAM2, PRKCH
|
|
Hypomethylation |
1 |
13 |
0.016389 |
ALB, BCHE, ADRA1A, AFP, GABRR3, PRL, ENAM, MAP2, PPIG, AMELX, HSD17B6, GAP43, CDK13
|
|
2 |
10 |
0.018602 |
IL5, IL18, TLR8, CCL3, CCL4, KLRK1, IL12RB2, SERPINB7, GPR115, TANK
|
References
1. Babu MS, Kaul S, Dadheech S, Rajeshwar K, Jyothy A, Munshi A : Serum albumin levels in ischemic stroke and its subtypes: correlation with clinical outcome. Nutrition 29 : 872-875, 2013   2. Chidambaran V, Zhang X, Martin LJ, Ding L, Weirauch MT, Geisler K, et al : DNA methylation at the mu-1 opioid receptor gene (OPRM1) promoter predicts preoperative, acute, and chronic postsurgical pain after spine fusion. Pharmgenomics Pers Med 10 : 157-168, 2017   3. Dorsch NW, King MT : A review of cerebral vasospasm in aneurysmal subarachnoid haemorrhage part I: incidence and effects. J Clin Neurosci 1 : 19-26, 1994   4. Duan W, Pan Y, Wang C, Wang Y, Zhao X, Wang Y, et al : Risk factors and clinical impact of delayed cerebral ischemia after aneurysmal subarachnoid hemorrhage: analysis from the China National Stroke Registry. Neuroepidemiology 50 : 128-136, 2018   5. Endres M, Meisel A, Biniszkiewicz D, Namura S, Prass K, Ruscher K, et al : DNA methyltransferase contributes to delayed ischemic brain injury. J Neurosci 20 : 3175-3181, 2000    6. Francoeur CL, Mayer SA : Management of delayed cerebral ischemia after subarachnoid hemorrhage. Crit Care 20 : 277, 2016    7. Guinot A, Lehmann H, Wild PJ, Frew IJ : Combined deletion of Vhl, Trp53 and Kif3a causes cystic and neoplastic renal lesions. J Pathol 239 : 365-373, 2016   8. Hamzei Moqaddam A, Ahmadi Musavi SM, Khademizadeh K : Relationship of opium dependency and stroke. Addict Health 1 : 6-10, 2009   9. Herman JG, Latif F, Weng Y, Lerman MI, Zbar B, Liu S, et al : Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc Natl Acad Sci U S A 91 : 9700-9704, 1994    10. Ieda M, Miyaoka T, Kawano K, Wake R, Inagaki T, Horiguchi J : May salivary alpha-amylase level be a useful tool for assessment of the severity of schizophrenia and evaluation of therapy? A case report. Case Rep Psychiatry 2012 : 747104, 2012    11. Kim BJ, Kim Y, Hong EP, Jeon JP, Yang JS, Choi HJ, et al : Correlation between altered DNA methylation of intergenic regions of ITPR3 and development of delayed cerebral ischemia in patients with subarachnoid hemorrhage. World Neurosurg 130 : e449-e456, 2019   12. Kim BJ, Kim Y, Kim SE, Jeon JP : Study of correlation between Hp α1 expression of haptoglobin 2-1 and clinical course in aneurysmal subarachnoid hemorrhage. World Neurosurg 117 : e221-e227, 2018   13. Kim BJ, Kim Y, Youn DH, Park JJ, Rhim JK, Kim HC, et al : Genome-wide blood DNA methylation analysis in patients with delayed cerebral ischemia after subarachnoid hemorrhage. Sci Rep 10 : 11419, 2020    14. Kim H, Crago E, Kim M, Sherwood P, Conley Y, Poloyac S, et al : Cerebral vasospasm after sub-arachnoid hemorrhage as a clinical predictor and phenotype for genetic association study. Int J Stroke 8 : 620-625, 2013   15. Knutsson A, Björkbacka H, Dunér P, Engström G, Binder CJ, Nilsson AH, et al : Associations of interleukin-5 with plaque development and cardiovascular events. JACC Basic Transl Sci 4 : 891-902, 2019    16. Kovacic MB, Myers JM, Wang N, Martin LJ, Lindsey M, Ericksen MB, et al : Identification of KIF3A as a novel candidate gene for childhood asthma using RNA expression and population allelic frequencies differences. PLoS One 6 : e237142011    17. Kurmi K, Haigis MC : Nitrogen metabolism in cancer and immunity. Trends Cell Biol 30 : 408-424, 2020    18. Kyadav K, Chaudhary HR, Gupta RC, Jain R, Yadav SR, Sharma S, et al : Clinical profile and outcome of stroke in relation to glycaemic status of patients. J Indian Med Assoc 102 : 138-139, 142, 1502004  19. Lad SP, Hegen H, Gupta G, Deisenhammer F, Steinberg GK : Proteomic biomarker discovery in cerebrospinal fluid for cerebral vasospasm following subarachnoid hemorrhage. J Stroke Cerebrovasc Dis 21 : 30-41, 2012   20. Lei L, Mason S, Liu D, Huang Y, Marks C, Hickey R, et al : Hypoxiainducible factor-dependent degeneration, failure, and malignant transformation of the heart in the absence of the von Hippel-Lindau protein. Mol Cell Biol 28 : 3790-3803, 2008    21. Li X, Lin S, Chen X, Huang W, Li Q, Zhang H, et al : The Prognostic value of serum cytokines in patients with acute ischemic stroke. Aging Dis 10 : 544-556, 2019    22. Liu D, Zempleni J : Transcriptional regulation of the albumin gene depends on the removal of histone methylation marks by the FAD-dependent monoamine oxidase lysine-specific demethylase 1 in HepG2 human hepatocarcinoma cells. J Nutr 144 : 997-1001, 2014    23. Liu Y, Gu HY, Zhu J, Niu YM, Zhang C, Guo GL : Identification of Hub genes and key pathways associated with bipolar disorder based on weighted gene co-expression network analysis. Front Physiol 10 : 1081, 2019    24. Martynov IuS, Shuvakhina NA : Carbohydrate metabolism disorders in stroke. Zh Nevropatol Psikhiatr Im S S Korsakova 79 : 1281-1288, 1979  25. McRonald FE, Morris MR, Gentle D, Winchester L, Baban D, Ragoussis J, et al : CpG methylation profiling in VHL related and VHL unrelated renal cell carcinoma. Mol Cancer 8 : 31, 2009    26. Nikkola E, Laiwalla A, Ko A, Alvarez M, Connolly M, Ooi YC, et al : Remote ischemic conditioning alters methylation and expression of cell cycle genes in aneurysmal subarachnoid hemorrhage. Stroke 46 : 2445-2451, 2015    27. Ott MO, Sperling L, Cassio D, Levilliers J, Sala-Trepat J, Weiss MC : Undermethylation at the 5' end of the albumin gene is necessary but not sufficient for albumin production by rat hepatoma cells in culture. Cell 30 : 825-833, 1982   28. Pascual JM, Carceller F, Roda JM, Cerdán S : Glutamate, glutamine, and GABA as substrates for the neuronal and glial compartments after focal cerebral ischemia in rats. Stroke 29 : 1048-1056; discussion 1056-1057, 1998   29. Peyravian N, Dikici E, Deo S, Toborek M, Daunert S : Opioid antagonists as potential therapeutics for ischemic stroke. Prog Neurobiol 182 : 101679, 2019    30. Qureshi WT, O'Neal WT, Khodneva Y, Judd S, Safford MM, Muntner P, et al : Association between opioid use and atrial fibrillation : the reasons for geographic and racial differences in stroke (REGARDS) study. JAMA Intern Med 175 : 1058-1060, 2015   31. Rowland MJ, Hadjipavlou G, Kelly M, Westbrook J, Pattinson KT : Delayed cerebral ischaemia after subarachnoid haemorrhage: looking beyond vasospasm. Br J Anaesth 109 : 315-329, 2012   32. Samuelsson C, Howells T, Kumlien E, Enblad P, Hillered L, Ronne-Engström E : Relationship between intracranial hemodynamics and microdialysis markers of energy metabolism and glutamate-glutamine turnover in patients with subarachnoid hemorrhage. Clinical article. J Neurosurg 111 : 910-915, 2009   33. Sang L, Wang XM, Xu DY, Zhao WJ : Bioinformatics analysis of aberrantly methylated-differentially expressed genes and pathways in hepatocellular carcinoma. World J Gastroenterol 24 : 2605-2616, 2018    34. Stamatovic SM, Phillips CM, Martinez-Revollar G, Keep RF, Andjelkovic AV : Involvement of epigenetic mechanisms and non-coding RNAs in blood-brain barrier and neurovascular unit injury and recovery after stroke. Front Neurosci 13 : 864, 2019    35. Starke RM, Kim GH, Komotar RJ, Hickman ZL, Black EM, Rosales MB, et al : Endothelial nitric oxide synthase gene single-nucleotide polymorphism predicts cerebral vasospasm after aneurysmal subarachnoid hemorrhage. J Cereb Blood Flow Metab 28 : 1204-1211, 2008   36. Suarez JI, Martin RH, Calvillo E, Dillon C, Bershad EM, Macdonald RL, et al : The albumin in subarachnoid hemorrhage (ALISAH) multicenter pilot clinical trial: safety and neurologic outcomes. Stroke 43 : 683-690, 2012    37. Zaina S, Heyn H, Carmona FJ, Varol N, Sayols S, Condom E, et al : DNA methylation map of human atherosclerosis. Circ Cardiovasc Genet 7 : 692-700, 2014   38. Zhao W, Lei T, Li H, Sun D, Mo X, Wang Z, et al : Macrophage-specific overexpression of interleukin-5 attenuates atherosclerosis in LDL receptor-deficient mice. Gene Ther 22 : 645-652, 2015  
|
|



















