Choi, Yun, Lee, Park, Park, and Kim: Proteomic Analysis of a Rat Cerebral Ischemic Injury Model after Human Cerebral Endothelial Cell Transplantation
Abstract
Objective
Cerebral endothelial cells have unique biological features and are fascinating candidate cells for stroke therapy.
Methods
In order to understand the molecular mechanisms of human cerebral endothelial cell (hCMEC/D3) transplantation in a rat stroke model, we performed proteomic analysis using 2-dimensional electrophoresis and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Protein expression was confirmed by quantitative real-time PCR and Western blot.
Results
Several protein spots were identified by gel electrophoresis in the sham, cerebral ischemia (CI), and CI with hCMEC/D3 treatment cerebral ischemia with cell transplantation (CT) groups, and we identified 14 differentially expressed proteins in the CT group. Proteins involved in mitochondrial dysfunction (paraplegin matrix AAA peptidase subunit, SPG7), neuroinflammation (peroxiredoxin 6, PRDX6), and neuronal death (zinc finger protein 90, ZFP90) were markedly reduced in the CT group compared with the CI group. The expression of chloride intracellular channel 4 proteins involved in post-ischemic vasculogenesis was significantly decreased in the CI group but comparable to sham in the CT group.
Conclusion
These results contribute to our understanding of the early phase processes that follow cerebral endothelial cell treatment in CI. Moreover, some of the identified proteins may present promising new targets for stroke therapy.
Key Words: Brain · Ischemia · Cell therapy · Proteomics · 2-dimensional electrophoresis.
INTRODUCTION
Stroke is the third most common cause of death in Korea 18). Interest in improving functional recovery after stroke is long-standing, and various therapeutic modalities, including exogenous cell therapy, have been applied to restore brain function 29). Exogenous cell therapy has shown promise as a means for augmenting brain repair, as evidenced by enhanced endogenous angiogenesis, neurogenesis, and immunomodulation 17). Neural stem cells and mesenchymal stem cells, which originate from a variety of sources, have been used for treatment in a focal cerebral ischemic animal model 8,36) and in human stroke patients 33). Cerebral endothelial cells have unique features compared to human umbilical vein endothelial cells (HUVEC) or endothelial precursor cells 30) and play an important role in maintaining the function of neurovascular units and promoting endogenous neural stem cell proliferation 11). The potential therapeutic benefits of human cerebral endothelial cells (HEN6) applied directly to the striatum of experimental ischemic stroke rats have been demonstrated 15). Recently, we reported that systemically injected human immortalized cerebral endothelial cells (hCMEC/D3) in a focal cerebral ischemia (CI) model successfully migrated into the ischemic hemisphere and had several neuroprotective effects, including reduced infarct size, neuroinflammation, and blood-brain barrier (BBB) breakdown, and enhanced functional recovery 7,25). Understanding the molecular alterations in the cell therapy model for animal stroke is critical for optimal translational strategy development. Very few studies have reported global protein expression changes in the human or rat ischemic brain 3,69). Monitoring following various kinds of cell therapy uses two-dimensional (2-D) electrophoresis-based protein expression analysis 12,1422). After the 2-D electrophoresis, distinct messenger ribonucleic acid (mRNA)-protein expression have evaluated by quantitative methods 19,24). Although neuroprotective events by transplanted hCMEC/D3 are usually observed around 3 days following cell therapy 7,25), the potential mechanisms of action are not completely understood. The aim of the present study was to compare the protein expression pattern in focal CI model rats treated with human cerebral endothelial cells in order to find new and robust potential therapeutic targets for stroke.
MATERIALS AND METHODS
Cells and reagents
The human cerebral microvascular endothelial cell line (hC-MEC/D3) was obtained from Merck Millipore (SCC066, Temecula, CA, USA) and cultured in an Endothelial Growth Medium-2 BulletKit (Lonza, Walkersville, MD, USA) as in our previous report 25). Primary antibodies against chloride intracellular channel 4 (CLIC4, Abcam, Cambridge, UK), paraplegin matrix AAA peptidase subunit (SPG7, OriGene, Rockville, MD, USA), peroxiredoxin 6 (PRDX6, Abcam, Cambridge, UK), zinc finger protein 90 (ZFP90, NovoPro, Shanghail, China), Eph receptor B1 (EPHB1, R&D Systems, Minneapolis, MN, USA), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH, Cell Signaling, Donvers, MA, USA) were used. Horseradish peroxidase (HRP)-conjugated secondary antibody was obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA). All chemicals used were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Animal model and cell transplantation
All surgical procedures and postoperative care in this study were performed in accordance with the guidelines of the Chonnam National University Animal Care and Use Committee. A total of 33 male Sprague-Dawley rats (8 weeks old) weighing 250-300 g were maintained on a 12 h light/dark cycle. Focal cortical ischemia was induced by photothrombosis of the cortical microvessels as described in our previous reports 7,25). Rats were randomly assigned to three groups : phosphate-buffered saline (PBS)-treated (sham), focal CI, and cell transplantion after focal CI (CT). A single cell suspension of 1×10 6 hCMEC/D3 cells in 10 µL of saline was injected into the left common carotid artery with a custom Hamilton syringe with a 33-gauge needle as in our previous report 25). Successful lesion making was confirmed by sharply departed cortical lesion in the motor cortex after open craniectomy. All brain samples for mRNA and protein expression analysis were obtained 3 days after focal CI induction.
Protein extraction
Following sacrifice (n=3, each group), tissue samples were isolated, weighed, mixed in three volumes of protein extraction buffer (8 M urea, 2 M thiourea), and homogenized. After centrifugation (8000 rpm, 30 min), the supernatant was collected and mixed with the same volume of 20% TCA/acetone to precipitate the proteins (-20℃, 1 h). The TCA was washed away with cold acetone (3 times), and the proteins were collected by centrifugation (10000 rpm, 15 min, 4℃). The precipitate was dried, dissolved in 200 µL protein extraction buffer, and stored at -70℃. Protein quantification was conducted using the BCA method with bovine serum albumin and confirmed with Western blotting against GAPDH.
Two-dimensional gel electrophoresis (2-DE)
Proteins were separated using 2D-PAGE and analyzed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry as described previously 10). The extracted proteins were solubilized with 200 µL of rehydration buffer containing 8 M urea, 2% CHAPS, 2% IPG buffer, 100 mM dithiothretiol (DTT), and 0.001% bromophenol blue. Then, 250 µL of rehydration buffer containing 1 mg of protein was applied to an immobilized pH 4-7 (NL) gradient (IPG) dry strip (13 cm; GE Healthcare, Amersham, UK), and the IPG strip was rehydrated for 12 h at 20℃. After rehydration, proteins were focused by isoelectric focusing (IEF). The voltage used was 300 V for 1 h, 3500 V for 90 min, and 3500 V for 4 h. The current was limited to 2 mA per strip. Following IEF, the resulting strip was equilibrated for reduction of proteins with equilibration buffer (50 mM Tris-Cl pH 8.8, 6 M urea, 30% glycerol, 2% sodium dodecyl sulfate (SDS), 0.001% bromophenol blue, and 1% DTT) for 20 min at room temperature and then incubated for protein alkylation with equilibration buffer containing 2.5% iodoacetamide but not DTT in the dark for 20 min at room temperature. The strips were separated on a 12% gel by the second dimension of electrophoresis and run in 45 mA at 20℃. To ensure reproducibility in the gel pattern, we repeated 2-DE against the same sample three times.
Gel staining and detection of proteins
The resulting gel was fixed in 12% trichloroacetic acid for 1 h and stained with Coomassie Blue G250 according to the colloidal Coomassie staining method. To identify the spots of interest, gels were excised and transferred into a microcentrifuge tube and then rinsed with distilled water for 15 min. Fifty percent acetonitrile (ACN) was added to the tube and incubated for 15 min. On discarding the ACN, 100% ACN was added to the tube and allowed to stand for 15 min until gel pieces became white and had shrunk. After removal of ACN, the gel was incubated with 100 mM ammonium bicarbonate (ambic) for 5 min. One hundred percent ACN was added to the tube and incubated for 15 min. This procedure was repeated until dye was no longer visible. Destained gel pieces were dried using a Speed Vac. Dried gel pieces can be stored at -70℃ prior to use.
Trypsin digestion and mass spectrometry
Destained gel pieces were treated with 0.1% (w/v) RapiGest SF in 50 mM ambic buffer for 10 min at 37℃ and then dried using a Speed Vac. Gel pieces were treated with 30 µL of trypsin solution (10 µg/mL) and incubated for 45 min on ice. After discarding excess trypsin solution, 30 µL of hydrolysis buffer (50 mM ambic/1 mM CaCl2) was added to the tube containing the gel piece. Finally, the gel pieces were incubated overnight at 37℃. Tryptic peptides were transferred into a new microcentrifuge tube and 20 µL of 50 mM ambic buffer was added to the tube containing the gel piece for 15 min. Then, 20 µL of ACN was added to the tube containing the gel piece and incubated for 15 min, and 40 µL of the eluted peptides was collected into the new tube containing tryptic peptides. Gel pieces were also incubated with 20 µL of 5% formic acid for 15 min, and 20 µL of ACN was added to the gel tube. The 5% formic acid/ACN (ratio 1 : 1) step was repeated to elute tryptic peptides. After adding 100 mM DTT to the tryptic peptides to a final concentration of 1 mM, the peptides were dried by using a vacuum centrifuge and stored at -70℃ for further analysis. The dried peptides were dissolved with 3 µL of buffer [50% ACN/0.1% trifluoroacetic acid (TFA)]. One microliter of peptide solution was mixed with 1 µL of α-cycano-4-hydroxycinnamic acid (CHCA) matrix solution (10 mg/mL of CHCA, 50% ACN, 49.9% EtOH, and 0.1% TFA), and then 1 µL of the sample/matrix mixture was directly deposited onto the plate and dried. Protein identification by peptide mass fingerprinting (PMF) was carried out using the MASCOT database (matrixscience.com). The parameters for PMF were as follows : mass error tolerance, 100 ppm; variable modification, oxidation of methionine and carbamidomethylation of cysteine; and the number of missed cleavages, 1. The mass data were calibrated with Angiotensin I (m/z 1296.6853) as an external standard and a peptide derived from trypsin (m/z of 2211.1046) as an internal standard.
Quantitative real-time PCR
Quantitative real-time PCR (qRT-PCR) was performed as described previously 7,25). The ribonucleic acid (RNA) of the ischemic hemisphere (n=5, each group) was extracted with Trizol reagent (Invitrogen, Carlsbad, CA, USA) and converted into complementary deoxyribonucleic acid (cDNA) by use of the GoScript™ cDNA Synthesis Kit (Promega Corporation, Madison, WI, USA) according to the manufacturer's instructions. The qRT-PCR was run on a QuantStudio 3 (ThermoFisher Scientific Inc., Waltham, MA, USA) in the presence of a florescent dye (SYBR™ Green master mix, ThermoFisher Scientific Inc., Waltham, MA, USA). The relative level of mRNA was calculated with the delta-delta cycle threshold method and presented after normalization to GAPDH. All primers were designed for Rattus norvegicus-derived sequences, and synthesized by Macrogen Inc. (Seoul, Korea), and the primer sequences used in this study for qRT-PCR were as follows : rat Clic4 : sense, 5'-ACATCTGCAGAACTTGGCTCCT-3', antisense, 5'-GTACGCAGAGAACTTGGCAAAGA-3'; rat Spg7 : sense, 5'-AGCTTGCTATTGCGAATACTGAC-3', antisense, 5'-CTTTGCCTTTGGGTTTATCATC-3'; rat Prdx6 : sense, 5'-CTTATCGTCGATGATGGGAAATGGT-3', antisense, 5'-GCCAGAGTTTGCCAAGAGGAATGT-3'; rat Zfp90 : sense, 5'-AGGGTTTCTCTCCGCTATGTGTGA-3', antisense, 5'-GCCTTCCAGCGCAGTTCATCT-3'; rat Ephb1 : sense, 5'-GTGGGCGAGATGATGTGACCTACA-3', antisense, 5'-ATCGCAGCGGGAGCAACTC-3'; and rat Gapdh : sense, 5'-GGGCATCCTGGGCTACACTGA-3', antisense, 5'-CCTTGCTGGGCTGGGTGGT-3'.
Western blot analysis
Western blot analysis was performed as previously described with proper modification 25). Briefly, ischemic hemispheres (n=4, each group) were lysed by the PRO-PREP™ Protein Extraction Kit (iNtRON Biotechnology, Seoul, Korea). After separation by 8-10% SDS-PAGE, proteins were transferred to polyvinylidene difluoride membrane and then incubated with primary antibody at 4℃ overnight. Each primary antibody described in the reagents section was used at 1 : 1000 dilution, except SPG7 (1 : 2000 dilution). GAPDH was used as the control. The membranes were washed with Phosphate Buffened Saline with Tween® 20 three times and further incubated with host-matched HRP-labeled secondary antibodies for 1 h at room temperature (1 : 1000; Santa Cruz). The Western blots were visualized using an enhanced chemiluminescence kit (ECL; GE Healthcare Life Sciences, Little Chalfont, Buckinghamshire, UK), and the optical densities were measured using a Fujifilm LAS-3000 luminescent image analyzer (FUJIFILM Corporation, Tokyo, Japan).
Statistical analysis
The statistical analyses were performed with PRISM 5.0 for Windows (GraphPad Software, Inc., La Jolla, CA, USA). Student's t-tests were used for comparisons between two groups, and analyses of variance and Bonferroni post hoc tests were used for comparisons among multiple groups. p-value of less than 0.05 was considered statistically significant.
RESULTS
2-D electrophoresis spot screening and proteomic analysis
The spot distribution patterns for the sham group, photothrombotic focal C) group, and CI with hCMEC/D3 cell transplantation (CT) group were screened (n=3, each group) 3 days after injury. The spot numbers of the sham and CT groups were detected between 754±83, but the spot numbers of the CI group were markedly increased (986±105) ( Fig. 1). Relative to the sham group, the intensity of 62 spots was significantly increased, and the intensity of 35 spots was significantly decreased in the CI group. Relative to the CI group, the intensity of 24 spots was increased, and the intensity of 32 spots was decreased in the CT group. After Coomassie Blue staining of resulting gels, 14 spots were sufficient for protein identification, carried out using the MASCOT database (matrixscience.com), by PMF. Fourteen spots are listed in Table 1. Among the 14 identified proteins, 12 proteins were downregulated in the CT group compared with those in the CI group, and 2 proteins in the CT group were recovered to sham group level.
qRT-PCR and Western blot analysis for identified target proteins
To confirm the results from the 2-DE analysis, mRNA expression levels were evaluated for the 14 identified proteins using qRT-PCR (n=5, each group) ( Fig. 2). Among them, Zfp90, Prdx6, Spg7, and Clic4 gene mRNA expression levels were statistically different between CI and CT groups ( p<0.01, Zfp90 and Clic4; p<0.05, Prdx6 and Spg7). Ephb1 mRNA expression level in the CT group was reduced compared with that in the CI group but did not reach statistical significance ( p=0.062). Finally, we performed Western blots to characterize the protein expression levels that showed differential mRNA expression between CI and CT groups (n=5, each group) ( Fig. 3). For the validation, we used the same rat brain samples that we used in the 2-DE analysis. The protein expression levels of EPHB1, ZFP90, and SPG7 were significantly downregulated and CLIC4 was significantly upregulated in the CT group compared with the CI group ( p<0.05, EPHB1; p<0.01, ZFP90, PRDX6, SPG7, and CLIC4).
DISCUSSION
In this study, we used 2-DE-based proteomics to elucidate the early phase neuroprotective mechanisms of human cerebral endothelial cell transplantation in focal CI. In the spot analysis, we identified 14 sequence-matched proteins based on the protein database, and using qRT-PCR and immunoblot analyses, we confirmed several protein expression changes after hCMEC/D3 transplantation in the early period of rat focal CI.
During CI, various detrimental processes occur, including overproduction of oxidants, inactivation of detoxification systems, and consumption of antioxidants. Mitochondria are known to play a key role in ischemic/reperfusion injury in the brain 16). Increased synthesis of TNF-α aggravates neuronal death by promoting mitochondrial dysfunction, calcium dysregulation, and oxidative stress 28). In our previous report, the hCMEC/D3 transplantation model decreased infarct size and attenuated TNF-α expression at day 3 after focal CI induction 25). In the present study, SPG7 expression is increased in the CI group at day 3 compared with sham expression, but in the CT group, expression is similar to sham level ( Fig. 3). SPG7 has recently become well known and is required for Ca 2+ overload- and ROS-induced permeability transition pore (PTP) opening, mitochondrial dysfunction, and cell death 1,31). In fact, abrupt SPG7 overexpression induces PTP opening after ischemic injury, suggesting that reduction of SPG7 expression could protect mitochondria from Ca 2+- and ROS-induced PTP-dependent necrosis 31). Our findings suggest that hCMEC/D3 application in the ischemic rat model may be neuroprotective by reducing TNF-α and SPG7 expression following CI. Brain ischemia induces PRDX6 and toll-like receptor 4 (TLR4) expression, which may contribute to the sequential neuroinflammation associated with CI 20). Interestingly, ligustilide may provide early and direct neuroprotection by inhibiting TLR4/PRDX6 signaling, suggesting that TLR4/PRDX6 blocking agents are potent neuroprotective tools 32). In the photothrombotic focal ischemia model, Prdx6 mRNA expression was markedly increased at day 3 and significantly reduced in the hCMEC/D3 treated group ( Fig. 2D), but the change in protein expression did not reach statistical significance ( Fig. 3B). These findings are consistent with our previous study that showed hCMEC/D3 transplantation can ameliorate production of neuroinflammatory cytokines, such as TNF-α and IL-1β, in the early phase of CI. The RE1-Silencing Transcription factor (REST), also known as Neuron-Restrictive Silencer Factor (NRSF), acts as a transcriptional repressor 27). ZFP90 is a novel NRSF-binding protein that inhibits the repressor activity of NRSF by preventing its binding to deoxyribonucleic acid 13,21). ZFP90 is highly expressed in pathological conditions such as congestive heart failure and aggravates cardiac dysfunction via increased expression of NRSF target genes, such as brain natriuretic peptide, present during adverse cardiac remodeling. However, the role of REST and ZFP90 in CI is only partially understood. In an in vitro model of ischemia, acute knockdown of REST in hippocampal slices subjected to oxygen glucose deprivation prevented GluA2 downregulation and neuronal death 4). In a stroke model, RNAi-mediated depletion of REST or administration of dominant-negative REST delivered directly into the hippocampus in vivo prevented epigenetic modifications, restored gene expression, and rescued hippocampal neurons 26). During CI/reperfusion injury, cerebrovascular maintenance functions are disrupted, and the goal of CI treatment is to restore vasculogenesis and neurogenesis, focusing on the ischemic penumbra. As our previous results and other studies have shown, cerebral endothelial cell transplantation plays a potent neurorestorative role, which involves protection against BBB breakdown through increased tight junction proteins 7) and promotion of vasculogenesis via vascular endothelial growth factor (VEGF) signaling 15). In the present study, we identified changes in CLIC4 expression using the 2-DE method; while CLIC4 is significantly decreased in the CI group, it is normalized relative to the sham group level in the hCMEC/D3 transplantation group. CLIC4 is normally expressed in endothelial cells 34) and vascular smooth muscle cells 35) and is essential for normal vasculogenesis through endothelial tubulogenesis during embryogenesis and ischemia 2). Clic4 gene deficiency results in reduced expression of Vegfr2, Vegfr1, Vegfc, and mural cell markers associated with a reduction in ischemia-induced expression of hypoxia inducible factor-1α (HIF-1α) 5). Recent work showed that overexpression of VEGF-A in Clic4 (-/-) mice rescued defects in collateral maturation by preventing mural cell loss and collateral pruning, thus restoring collateral number and diameter and reducing stroke severity in adults 23). In fact, hCMEC/D3 showed high Clic4 mRNA and protein expression, whereas HUVEC did not (data not shown). Considering previous reports and the present study, transplanted Clic4-expressing hC-MEC/D3 cells could migrate to the ischemic lesion 25) and contribute to microenvironmental improvement by restoring the HIF-1α/VEGF axis. In this study, focal CI induced various protein expression changes associated with promoting apoptosis and neuronal cell death and repressing vasculogenesis. Transplanted cerebral endothelial cells (hCMEC/D3) may partly play a role in controlling neuroinflammation via SPG7 and PRDX6 suppression, reducing fetal gene re-expression via ZFP90, and modulating the damaged vasculature via CLIC4 upregulation compared with the CI group. And we could assume that these effects are strongly corroborates the paracrine mediated neuroprotection of hCMEC/D3 cells on ischemic damaged hemisphere 17). However, our study is limited since we only focused on the events at day 3 following focal CI. Because neuronal plasticity is combined with angiogenesis and neurogenesis after severe injury, studies that show chronological target protein changes in hC-MEC/D3-treated animals will provide meaningful information on the neurorestorative mechanisms of cerebral endothelial cell therapy.
CONCLUSION
Human cerebral endothelial cell transplantation following focal CI induced several protein expression changes. Aberrantly increased expression of proteins associated with neuroinflammation or apoptosis, such as ZFP90, PRDX6, and SPG7, was decreased following cell transplantation. CLIC4 expression in the cell transplantation group was recovered to sham expression level. These results contribute to our understanding of the early phase processes that follow cerebral endothelial cell treatment in CI. Moreover, some of the identified proteins may present promising new targets for stroke therapy.
Acknowledgements
This study was supported by a grant (HCRI 16921-21) from the Chonnam National University Hwasun Hospital Institute for Biomedical Science.
References
1. Bernardi P, Forte M : Commentary : SPG7 is an essential and conserved component of the mitochondrial permeability transition pore. Front Physiol 2015, 6 : 320,    2. Bohman S, Matsumoto T, Suh K, Dimberg A, Jakobsson L, Yuspa S, et al : Proteomic analysis of vascular endothelial growth factor-induced endothelial cell differentiation reveals a role for chloride intracellular channel 4 (CLIC4) in tubular morphogenesis. J Biol Chem 2005, 280 : 42397-42404,   3. Brea D, Agulla J, Staes A, Gevaert K, Campos F, Sobrino T, et al : Study of protein expression in peri-infarct tissue after cerebral ischemia. Sci Rep 2015, 5 : 12030,    4. Calderone A, Jover T, Noh KM, Tanaka H, Yokota H, Lin Y, et al : Ischemic insults derepress the gene silencer REST in neurons destined to die. J Neurosci 2003, 23 : 2112-2121,    5. Chalothorn D, Zhang H, Smith JE, Edwards JC, Faber JE : Chloride intracellular channel-4 is a determinant of native collateral formation in skeletal muscle and brain. Circ Res 2009, 105 : 89-98,    6. Chen JH, Kuo HC, Lee KF, Tsai TH : Global proteomic analysis of brain tissues in transient ischemia brain damage in rats. Int J Mol Sci 2015, 16 : 11873-11891,    7. Choi KH, Kim HS, Park MS, Kim JT, Kim JH, Cho KA, et al : Regulation of caveolin-1 expression determines early brain edema after experimental focal cerebral ischemia. Stroke 2016, 47 : 1336-1343,   8. Cohen LK, Jensen MB : Scaffolds for intracerebral grafting of neural progenitor cells after cerebral infarction : a systematic review. Arch Neurosci 2015, 2 : e25364,    9. Cuadrado E, Rosell A, Colomé N, Hernández-Guillamon M, García-Berrocoso T, Ribo M, et al : The proteome of human brain after ischemic stroke. J Neuropathol Exp Neurol 2010, 69 : 1105-1115,   10. Granvogl B, Plöscher M, Eichacker LA : Sample preparation by in-gel digestion for mass spectrometry-based proteomics. Anal Bioanal Chem 2007, 389 : 991-1002,   11. Guo Y, Shi D, Li W, Liang C, Wang H, Ye Z, et al : Effects of cerebral microvascular endothelial cells and vascular endothelial growth factor on the proliferation and differentiation of NSCs : a comparative study. Br J Neurosurg 2010, 24 : 62-68,   12. Harvey A, Yen TY, Aizman I, Tate C, Case C : Proteomic analysis of the extracellular matrix produced by mesenchymal stromal cells : implications for cell therapy mechanism. PLoS One 2013, 8 : e79283,    13. Hata L, Murakami M, Kuwahara K, Nakagawa Y, Kinoshita H, Usami S, et al : Zinc-finger protein 90 negatively regulates neuron-restrictive silencer factor-mediated transcriptional repression of fetal cardiac genes. J Mol Cell Cardiol 2011, 50 : 972-981,   14. Dongsheng H, Zhuo Z, Jiamin L, Hailan M, Lijuan H, Fan C, et al : Proteomic analysis of the peri-infarct area after human umbilical cord mesenchymal stem cell transplantation in experimental stroke. Aging Dis 2016, 7 : 623-634,    15. Ishikawa H, Tajiri N, Shinozuka K, Vasconcellos J, Kaneko Y, Lee HJ, et al : Vasculogenesis in experimental stroke after human cerebral endothelial cell transplantation. Stroke 2013, 44 : 3473-3481,    16. Jackson MJ, Papa S, Bolaños J, Bruckdorfer R, Carlsen H, Elliott RM, et al : Antioxidants, reactive oxygen and nitrogen species, gene induction and mitochondrial function. Mol Aspects Med 2002, 23 : 209-285,   17. Kalladka D, Muir KW : Brain repair : cell therapy in stroke. Stem Cells Cloning 2014, 7 : 31-44,   18. Kim JS : Stroke becomes the 3rd important cause of death in Korea; is it a time to toast? J Stroke 2014, 16 : 55-56,    19. Koussounadis A, Langdon SP, Um IH, Harrison DJ, Smith VA : Relationship between differentially expressed mRNA and mRNA-protein correlations in a xenograft model system. Sci Rep 2015, 5 : 10775,    20. Kuang X, Wang LF, Yu L, Li YJ, Wang YN, He Q, et al : Ligustilide ameliorates neuroinflammation and brain injury in focal cerebral ischemia/reperfusion rats : involvement of inhibition of TLR4/peroxiredoxin 6 signaling. Free Radic Biol Med 2014, 71 : 165-175,   21. Kuwahara K, Saito Y, Ogawa E, Takahashi N, Nakagawa Y, Naruse Y, et al : The neuron-restrictive silencer element-neuron-restrictive silencer factor system regulates basal and endothelin 1-inducible atrial natriuretic peptide gene expression in ventricular myocytes. Mol Cell Biol 2001, 21 : 2085-2097,    22. Li WY, Jin RL, Hu XY, Chen W, Bang OY : Proteomic analysis of ischemic rat brain after human mesenchymal stem cell transplantation. Tissue Eng Regen Med 2014, 11 : 333-339,  23. Lucitti JL, Tarte NJ, Faber JE : Chloride intracellular channel 4 is required for maturation of the cerebral collateral circulation. Am J Physiol Heart Circ Physiol 2015, 309 : H1141-H1150,    24. Maier T, Güell M, Serrano L : Correlation of mRNA and protein in complex biological samples. FEBS Lett 2009, 583 : 3966-3973,   25. Moon JH, Na JY, Lee MC, Choi KH, Lee JK, Min JJ, et al : Neuroprotective effects of systemic cerebral endothelial cell transplantation in a rat model of cerebral ischemia. Am J Transl Res 2016, 8 : 2343-2353,   26. Noh KM, Hwang JY, Follenzi A, Athanasiadou R, Miyawaki T, Greally JM, et al : Repressor element-1 silencing transcription factor (REST)-dependent epigenetic remodeling is critical to ischemia-induced neuronal death. Proc Natl Acad Sci U S A 2012, 109 : E962-E971,    27. Ooi L, Wood IC : Chromatin crosstalk in development and disease : lessons from REST. Nat Rev Genet 2007, 8 : 544-554,   28. Pandya JD, Sullivan PG, Pettigrew LC : Focal cerebral ischemia and mitochondrial dysfunction in the TNFα-transgenic rat. Brain Res 2011, 1384 : 151-160,    29. Rodríguez-Frutos B, Otero-Ortega L, Gutiérrez-Fernández M, Fuentes B, Ramos-Cejudo J, Díez-Tejedor E : Stem cell therapy and administration routes after stroke. Transl Stroke Res 2016, 7 : 378-387,   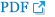 30. Schmidt NO, Koeder D, Messing M, Mueller FJ, Aboody KS, Kim SU, et al : Vascular endothelial growth factor-stimulated cerebral microvascular endothelial cells mediate the recruitment of neural stem cells to the neurovascular niche. Brain Res 2009, 1268 : 24-37,   31. Shanmughapriya S, Rajan S, Hoffman NE, Higgins AM, Tomar D, Nemani N, et al : SPG7 is an essential and conserved component of the mitochondrial permeability transition pore. Mol Cell 2015, 60 : 47-62,    32. Shichita T, Hasegawa E, Kimura A, Morita R, Sakaguchi R, Takada I, et al : Peroxiredoxin family proteins are key initiators of post-ischemic inflammation in the brain. Nat Med 2012, 18 : 911-917,   33. Steinberg GK, Kondziolka D, Wechsler LR, Lunsford LD, Coburn ML, Billigen JB, et al : Clinical outcomes of transplanted modified bone marrow-derived mesenchymal stem cells in stroke : a phase 1/2a study. Stroke 2016, 47 : 1817-1824,    34. Ulmasov B, Bruno J, Gordon N, Hartnett ME, Edwards JC : Chloride intracellular channel protein-4 functions in angiogenesis by supporting acidification of vacuoles along the intracellular tubulogenic pathway. Am J Pathol 2009, 174 : 1084-1096,    35. Wojciak-Stothard B, Abdul-Salam VB, Lao KH, Tsang H, Irwin DC, Lisk C, et al : Aberrant chloride intracellular channel 4 expression contributes to endothelial dysfunction in pulmonary arterial hypertension. Circulation 2014, 129 : 1770-1780,    36. Zhou F, Gao S, Wang L, Sun C, Chen L, Yuan P, et al : Human adipose-derived stem cells partially rescue the stroke syndromes by promoting spatial learning and memory in mouse middle cerebral artery occlusion model. Stem Cell Res Ther 2015, 6 : 92,   
Fig. 1
Representative two-dimensional gel electrophoretograms of rat brain tissue. (A) Sham group, (B) Photothrombotic focal cerebral ischemia (CI) group, and (C) CI treated with human cerebral endothelial cells (hCMEC/D3; CT) group. Circles show identified proteins that exhibit differences in average abundance values for each group (n=3 per group).
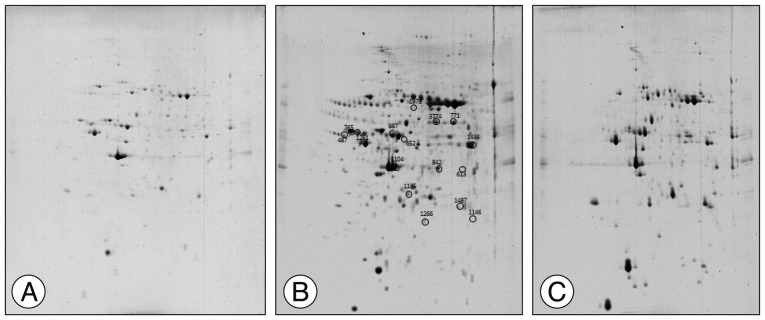
Fig. 2
qRT-PCR for selected target genes in the CT group compared with sham and CI groups. The mRNA levels of Ephb1, Zfp90, Prdx6, Spg7, and Clic4 were measured by qRT-PCR and calculated by the ΔΔCt methodology as described in methods. The assays were run in triplicate, and the standard error of the mean (SEM) was determined. Relative expression of Zfp90, Prdx6, and Spg7 were markedly decreased in the CT group compared with the CI group, but changes in Ephb1 did not reach statistical significance (A-D). mRNA for Clic4 was markedly decreased in the CI group but was rescued in the CT group (E; n=5 per group). Data are presented as mean±SEM. *p<0.05, **p<0.01 vs. CT group. qRT-PCR : quantitative real-time PCR, CI : cerebral ischemia, mRNA : messenger ribonucleic acid, EPHB1 : eph receptor B1, ZFP90 : zinc finger protein 90, PRDX6 : peroxiredoxin 6, SPG7 : paraplegin matrix AAA peptidase subunit, CLIC4 : chloride intracellular channel 4, GAPDH : glyceraldehyde 3-phosphate dehydrogenase.
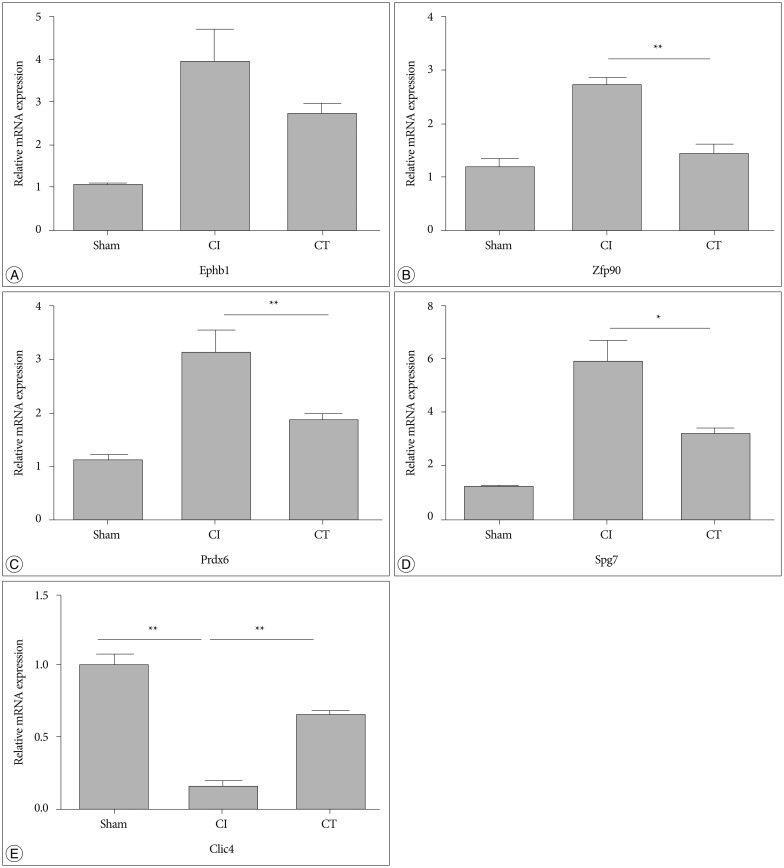
Fig. 3
Western blot analysis of proteomic and qRT-PCR results from CI and CT groups. A : Cropped bands of the Western blot gel corresponding to EPHB1, ZFP90, PRDX6, SPG7, and CLIC4 by group. B : Proteins that were increased in the CI group were markedly decreased in the CT group. C : Reduced CLIC4 in the CI group was normalized to sham level in the CT group (n= 3 per group). Data are presented as mean±SEM. *p<0.05, **p<0.01 vs. CT group. qRT-PCR : quantitative real-time PCR, CI : cerebral ischemia, EPHB1 : eph receptor B1, ZFP90 : zinc finger protein 90, PRDX6 : peroxiredoxin 6, SPG7 : paraplegin matrix AAA peptidase subunit, CLIC4 : chloride intracellular channel 4, SEM : standard error of the mean.
Table 1
List of identified differentially expressed proteins among three groups

|
|



















