Hong, Cho, Rhim, Park, Ahn, Youn, Kim, Park, Lee, Jeon, and on behalf of the First Korean Stroke Genetics Association Research (The FirstKSGAR) Study: Updated Trans-Ethnic Meta-Analysis of Associations between Inflammation-Related Genes and Intracranial Aneurysm
Abstract
Objective
We performed an expanded multi-ethnic meta-analysis to identify associations between inflammation-related loci with intracranial aneurysm (IA) susceptibility. This meta-analysis possesses increased statistical power as it is based on the most data ever evaluated.
Methods
We searched and reviewed relevant literature through electronic search engines up to August 2022. Overall estimates were calculated under the fixed- or random-effect models using pooled odds ratio (OR) and 95% confidence intervals (CIs). Subgroup analyses were performed according to ethnicity.
Results
Our meta-analysis enrolled 15 studies and involved 3070 patients and 5528 controls including European, Asian, Hispanic, and mixed ethnic populations. Of 17 inflammation-related variants, the rs1800796 locus (interleukin [IL]-6) showed the most significant genome-wide association with IA in East-Asian populations, including 1276 IA patients and 1322 controls (OR, 0.65; 95% CI, 0.56-0.75; p=3.24×10-9) under a fixed-effect model. However, this association was not observed in the European population (OR, 1.09; 95% CI, 0.80-1.47; p=0.5929). Three other variants, rs16944 (IL-1β), rs2195940 (IL-12B), and rs1800629 (tumor necrosis factor-α) showed a statistically nominal association with IA in both the overall, as well as East-Asian populations (0.01<p<0.05).
Conclusion
Our updated meta-analysis with increased statistical power highlights that rs1800796 which maps on the IL-6 gene is associated with IA, and in particular confers a protective effect against occurrence of IA in the East-Asian population.
Key Words: Inflammation mediators · Interleukin-6 · Intracranial aneurysm · Meta-analysis.
INTRODUCTION
Intracranial aneurysm (IA) refers to abnormal dilatation of the cerebral artery with symptoms ranging from asymptomatic, to wall rupture with life-threatening consequences. IA can usually be treated well by surgical clipping and endovascular coil embolization if the IA is detected before rupture. In contrast, the mortality and disability rates are quite high in the event of rupture as it results in subarachnoid hemorrhage (SAH) [ 15]. Accordingly, research in various fields from clinical trials to genetic research and artificial intelligence has been conducted to identify high-risk groups which may be predisposed to IA formation and rupture [ 1, 11, 15, 21]. Clinical risk factors for IA include smoking, hypertension, family history of IA, and female gender [ 7, 31]. In terms of pathophysiologic mechanisms, inflammation play a critical role in IA formation, growth, and rupture [ 5]. It is known that IA formation is primarily related to endothelial dysfunction, inflammatory responses, and a series of vascular smooth muscle cell abnormalities including phenotype change, proliferation, migration, and apoptosis [ 23]. These structural changes from pathological remodeling in the face of inflammation, result in loosening of the vascular wall integrity and aneurysmal dilatation [ 5, 23]. Infiltration of macrophages into the IA wall has been shown to correlate with degradation of extracellular matrix [ 20]. Jayaraman et al. [ 17] reported markedly elevated expression of tumor necrosis factor (TNF)-α in human IA, suggesting its potential role in inflammation-mediated promotion of, and subsequent apoptosis of the cerebral arteries. Mechanistically, the activity of the TNF-α converting enzyme responsible for TNF-α release, was induced after IA formation. Further, a series of inflammatory responses such as nuclear factor-κB activation, cyclooxygenase-2 (COX-2) expression, and macrophage infiltration into the IA wall, were significantly diminished in a mouse model of TNF receptor superfamily member 1a deficiency [ 2]. Also, interleukin (IL)-6 levels in blood samples taken from the IA orifice were closely associated with outcomes in patients with SAH [ 19]. Based on the results of studies like these, it appears it should be possible to characterize high-risk groups by analyzing the association of inflammation-related genes in relation to IA formation or rupture. Several inflammation-related gene loci have been reported to be associated with IA susceptibility using single nucleotide polymorphism (SNP) or genetic linkage analysis. Among the various inflammation-related genetic variants, a previous meta-analysis revealed that the rs1800629 polymorphisms (TNF-α) was closely associated with IA in Caucasians [ 16]. Also, rs1800796 (IL-6) showed opposite effects with respect to the formation of IA between Caucasian and Chinese populations, although there was no statistical significance. In 2020, Xu et al. [ 39] reported that rs1799964 (TNF-α) and rs17561 (IL-1A) were significantly associated with IA in dominant and additive models in the Chinese population. Recently, we reported the updated results of genome-wide association study by genotype correction and imputation in Korean patients with IA. As an extension of this study, we have now evaluated the association between inflammation-related loci and IA susceptibility via multi-ethnic meta-analysis with increased statistical power based on the most data thus far.
MATERIALS AND METHODS
All the study protocols have been approved by the Institutional Review Boards and Ethics Committees of the Hallym University Chuncheon Sacred Heart Hospital (No. 2017-9, 2018-6, and 2019-6). Informed consent was obtained from the patients or their relatives.
Literature review and study collection
We searched for English-language publications that investigated inflammation-related genes associated with IA patients. Electronic databases including PubMed, Embase, and Web of Science were used for articles that had been published up to August 2022. The following key words were determined by referring to previous articles and used in “and” combinations [ 16] : “intracranial aneurysm(s)” or “cerebral aneurysm(s)” or “subarachnoid hemorrhage”, “inflammation” or “inflammatory cytokine” or “interleukin” or “tumor necrosis factor” or “high mobility group box 1” or “genetic” ( Fig. 1 and Supplementary Table 1). We further included studies that contained the following criteria : 1) saccular aneurysm; 2) studies providing information of odds ratios (ORs), 95% confidence intervals (CIs), and allele frequency; and 3) case-control study. Exclusion criteria were : 1) non-saccular aneurysm such as fusiform or dissection; 2) non-spontaneous aneurysm due to infection or trauma; 3) review, comment, or case report; and 4) animal study. Data extraction were performed by two investigators (E.P.H. and D.H.Y.) independently. Disagreements were resolved by the third investigator (J.P.J.). Clinical information (e.g., authors, year, ethnicity, age, gender, and sample size) and genetic information (e.g., gene, SNP, and allele frequency) were collected. The quality of enrolled studies was evaluated using the Newcastle-Ottawa quality assessment scale [ 16]. The fixed-effect model assumes that one true effect size underlies all studies in the meta-analysis having a less heterogeneity between studies. This model is known to be suitable for a small number of studies. On the other hand, the random-effect model assumes that the effect size is not identical across the all studies involved in the meta-analysis. This approach can be suitable for a large number of studies to generalize the results beyond the included studies
Statistical analysis
Meta-analysis based on the inverse-variance method was performed using the Genome-Wide Association Meta-Analysis (GWAMA) software ( http://www.well.ox.ac.uk/gwama/) [ 25]. Overall estimates were calculated under the fixed- or randomeffect estimations. The Cochran Q test was performed for estimation of I 2 statistics to detect potential publication bias across individual studies. Funnel plots were illustrated for the visualization of a potential publication bias. Forest plots were drawn to describe effect size of each study and overall outcome. The plots were generated using the STATA software v.17.0 (Stata Corp., College Station, TX, USA).
RESULTS
Study characteristics of enrolled studies
After searching for and identifying an initial 808 articles, those that did not meet the inclusion criteria and duplicates were continuously removed. Finally, a total of 15 studies including 3070 patients and 5528 controls were included for meta-analysis ( Fig. 1). The enrolled studies included three ethnicities including six European-ancestry, seven Asian (six EastAsian and one South-Asian), one Hispanic Latin American, and one mixed population. Detailed demographic characteristics of the enrolled studies are presented in Table 1. With the exception of one study [ 40], the mean age of patients and controls in all others was over 40 years. One study had mixed-ethnic population including European-ancestry, African-American, and other ethnic subjects [ 12].
Table 2 shows summary statistics for the 17 total variants of inflammation-related genes such as IL-1α, IL-1β, IL-6, IL-12B, TNF-α, and high mobility group box 1 (HMGB1) in terms of variant name, effect allele frequency (EAF), genotype frequency, and OR with 95% CIs. Sixteen variants showed an association result by allelic or additive effect models in each study with the exception of one study [ 12] which used a dominant inherited test, not but not allelic or additive effect model test. Thus, the meta-analysis of the rs2249825 locus (HMGB1) was performed under the dominant model using the two studies [ 12, 13]. These two studies showed a same effect direction in the allele “G” of rs2249825, but a completely opposite frequency in that allele type (i.e., 0.150 vs. 0.887).
Meta-analysis of the inflammatory-related loci in the multi-ethnic populations
Estimates of the multi-ethnic meta-analysis of 17 variants under the fixed- or random-effect inverse variance models are presented in Table 3. In general, these variants did not pass a genome-wide significance threshold in the overall meta-analysis ( p<5×10 -8). The three inflammatory-gene loci of rs16944 (IL-1β), rs2195940 (IL-12B), and rs1800629 (TNF-α), showed a statistically nominal association in the overall fixed- and random-effect model ( p=0.0390, 0.0413, and 0.026, respectively) without heterogeneity across the studies. In particular, the rs1800796 (IL-6) variant showed a suggestive protective association in IA formation (OR, 0.75; 95% CI, 0.67-0.85; p=1.81×10 -6) in the overall fixed-effect model composed of 1886 IA patients and 4454 controls. After excluding three studies that caused heterogeneity [ 28, 37, 40], this variant still showed a marginal association with IA formation (OR, 0.87; 95% CI, 0.76-0.99; p=0.0455) ( Fig. 2). Except for rs1800796, other variants did not show an association with IA susceptibility. In the subgroup analysis, the EAF for rs1800796 exhibited a clear difference between European- and Asian-ancestry populations (EAF, 0.062 vs. 0.481). Interestingly, this locus was associated with protective effects on IA formation in the East-Asian subgroup meta-analysis (fixed-effect : OR, 0.65; 95% CI, 0.56-0.75; p=3.24×10 -9) but not the European population subgroup meta-analysis (fixed-effect : OR, 1.09; 95% CI, 0.80-1.47; p=0.5929). However, heterogeneity across the studies was observed ( Table 4). When the analysis was conducted including studies targeting East-South Asian populations, the rs1800796 showed a suggestive association with IA in the fixed-effect model (OR, 0.72; 95% CI, 0.62-0.80; p=7.28×10 -8), but not in the random-effect model. Inflammation-related variants with the exception of rs16944 (IL-1β), rs2195940 (IL-12B), and rs1800796 (IL-6), did not possess significant or marginal associations ( p-values of less than 0.05) with IA susceptibility in either European- or Asian-ancestry meta-analyses ( Table 4).
DISCUSSION
Although the exact mechanisms by which inflammation is involved in IA pathogenesis are still uncertain, it is generally known that inflammatory factors play an important role in the development, growth, and rupture of IA [ 27]. Among various inflammation-related variants, our study revealed that the rs1800796 (IL-6) showed the greatest overall significance with IA susceptibility (OR, 0.75; 95% CI, 0.67-0.85; p=1.81×10 -6) under a fixed-effect model. However, study heterogeneity across studies can be a concern. To address this issue, we performed the meta-analysis again after excluding studies with heterogeneity that significantly affected the results through funnel plot ( Fig. 2). The results showed that the pooled OR was 0.87 (95% CI, 0.76-0.99) under a fixed effect model with a heterogeneity of 34.13%, and still exhibiting the association between rs1800796 and IA. When performing subgroup analysis based on ethnicity, the association was more evident in the East-Asian populations than in the European population. Accordingly, the rs1800796 allele demonstrates an association with IA, which occurs particularly in Asians. IL-6 mediated inflammatory signaling results in IA formation via activation of monocyte chemoattractant protein-1, matrix metallopeptidase 9, T helper 17, and macrophage and monocyte infiltration [ 27]. Investigations into the role of IL-6 in IA formation and rupture has principally been performed in rodent models of IA. However, previous studies have shown somewhat conflicting results leading to uncertainty as to whether IL-6 is a risk factor or a protective factor for IA formation and rupture. Overexpression of micro ribonucleic acid (miR)-21 increased the risk of IA formation and rupture via stimulation of c-Jun N-terminal kinase signaling pathway-mediated inflammation, with increased IL-6 [ 6]. Kao et al. [ 19] reported that high levels of IL-6 at IA were closely associated with poor neurologic outcomes at 1 month, while high venous IL-6 was marginally associated with poor outcomes. On the other hand, IL-6 gene expression was significantly increased in IA induction in lymphocyte-deficient (Rag1) mice compared to wild-type mice, indicating a protective role of IL-6 in IA formation [ 34]. In terms of genetic predisposition to IA, IL-6 SNPs have been the main focus of investigation. McColgan et al. [ 26] reported that IL-6 gene G572C polymorphism exhibited a significant IA association. Further, IL-6 gene G174C polymorphism showed a protective effect against IA formation after analysis of three studies. In this meta-analysis, the C allele of rs1800796 (IL-6) exhibited a protective effect against IA formation while the G allele increased the risk of IA. Although not specifically IA, it was shown when analyzing the -174 G/C polymorphism (IL-6), that the GG genotype was a risk factor for patients with multi-infarct dementia compared with the CC genotype [ 32]. Therefore, it will be important to investigate what differences are present in the IA wall as a result of expression of different SNP alleles, through follow-up studies. In this study, three variants, namely, rs16944 (IL-1β), rs2195940 (IL-12B), and rs1800629 (TNF-α) showed a statistically nominal association with IA. Based on the observation that an IL-1β gene polymorphism was associated with severity of inflammatory bowel disease, studies on the role of IL-1β gene and aneurysm formation have been conducted [ 29, 35]. Aortas from IL-1β and IL-1R knockout mice revealed decreased inflammatory cytokine and matrix metalloproteinase 9 expression [ 18]. Slowik et al. [ 35] reported that the TT genotype of IL-1β 511 C/T polymorphisms (rs16944) significantly increased the risk of SAH in patients compared to controls. Our meta-analysis also showed that rs16944 (IL-1β) increased the risk of IA (OR, 1.13; 95% CI, 1.01-1.28; p=0.0390). IL-12B is located at chromosome 5q33.3 [ 16]. Li et al. [ 22] reported that the AC/CC genotypes and C allele of rs3212227 (IL-12B) increased the IA risk more than the AA genotype and A allele. In particular, the association was more evident in males than females. However, previous meta-analyses did not confirm the association between rs3212227 (IL-12B) polymorphisms and IA [ 16, 33]. We also did not find this association after analyzing four studies with fixed effect or random effect models. Meanwhile, rs2195940 (IL-12B) showed a significant protective effect on IA formation without heterogeneity (OR, 0.65; 95% CI, 0.43- 0.98; p=0.0413). TNF-α is a well-known pro-inflammatory cytokine. Starke et al. [ 36] reported that TNF-α knockout mice or mice pre-treated with a synthesized TNF-α inhibitor exhibited significantly lower incidence of IA formation and rupture than sham controls. Expression of TNF-α mRNA and protein were also elevated. Consistent with the results of the meta-analysis published in 2020 [ 16], we also observed a statistically nominal association, but not a genome-wide significance level (OR, 0.78; 95% CI, 0.62-0.97; p=0.0260). Nevertheless, the association between IA and differences in the allele or genetic frequency of the three variants as compared to their expression in tissue or blood, is not clear. Accordingly, future studies should address mechanistic differences between the three variant SNPs in IA formation and growth. In this study, we also analyzed HMGB1 SNPs in relation to IA. Hendrix et al. [ 12] reported that the G allele of rs2249825 increased the risk of delayed cerebral ischemia in patients with SAH. However, there was no significant difference in genotype frequencies associated with IA when compared to controls. We also observed that rs2249825 did not have a meaningful association with IA in the fixed effect model, although it increased IA risk the most among the inflammation-related variants we analyzed (OR, 1.47; 95% CI, 0.98-2.22; p=0.0645). However, only two studies were analyzed, and heterogeneity between those studies was high. Considering that HMGB1 is associated with vascular stiffness and inflammatory responses in endothelial cells, both of which are highly related to IA [ 30], a more comprehensive analysis, including HMGB1 gene variants and their expression of mRNA and protein is required. Although this study is an upgraded meta-analysis with the largest number of studies and subjects to date, it has some limitations. First, the results of this meta-analysis could be biased depending on clinical factors (e.g., age, gender, and underlying diseases) and the analytic methods (such as allelic or genetic model-based SNP association tests e.g., additive or dominant model) that are different for each study. Second, various inflammation-related SNP analyses have been performed, but the common SNPs studied in multiple studies and ethnicities are very limited and rs1800796 (IL-6) is representative. Third, the overall effect of study results showed a moderate or high heterogeneity between intra-population (e.g., within same nationality such as Chinese studies) and inter-population. Therefore, further meta-analysis including more studies that investigate inflammatory-related markers under the level of genetic or genome-wide associations are required. Furthermore, research should be conducted on whether SNPs which are known to be associated with IA via meta-analysis, can be verified in over 10K subjects in a large-scale population study. Finally, it is challenging to find missing heritability to the point variation since IA formation and growth are thought to be influenced by a variety of factors, including not only genetic factors but also clinical factors [ 14, 15]. In this regard, we emphasize that studies on the biological functions of variants associated with IA would be informative [ 38].
CONCLUSION
Our updated meta-analysis shows that rs1800796 (IL-6) gene is closely associated with IA, and in particular offers a protective effect against IA formation in the East-Asian population. As well, three additional inflammation-related variants (rs16944 [IL-1β], rs2195940 [IL-12B], and rs1800629 [TNF-α]) have a possible genetic association in IA formation in European and Asian populations.
Acknowledgements
This research was supported by a grant of the Hallym University Research Fund (HURF-2021-35). This work was supported by the Korea Bio Data Station (K-BDS) with computing resources including technical support from 2022 to 2023.
Fig. 1.
PRISMA flow diagram of candidate study selection from reviewing previous genetic or genome-wide association studies focused on inflammation-related genes in intracranial aneurysm. PRISMA : Preferred Reporting Items for Systematic Reviews and Meta-Analyses. 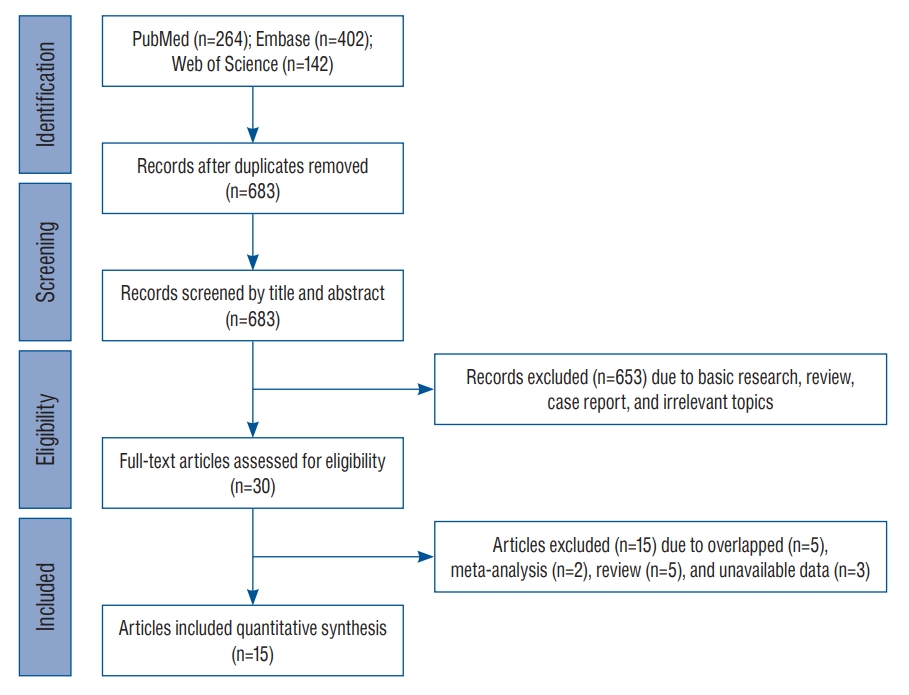
Fig. 2.
A : Funnel plot with pseudo 95% confidence limits for rs1800796 (interleukin [IL]-6) in the overall meta-analysis. X-axis indicates the natural logarithm of odds ratio (ln[OR]). Y-axis indicates standard error of ln(OR). Solid and short-dash lines indicate the overall effect and 95% confidence intervals, respectively. B : Forest plots of rs1800796 in the inverse variance meta-analysis excluding study heterogeneity. The diamonds indicate estimates of pooled OR from the fixed-effect (first diamond) and random-effect (second diamond) models. L95 : lower 95% confidence interval, U95 : upper 95% confidence interval. 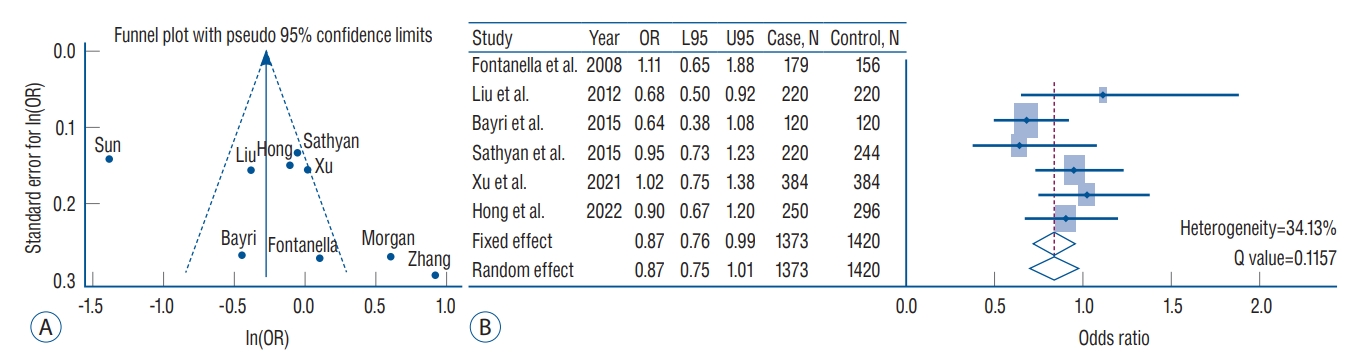
Table 1.
Clinical characteristics of the thirteen studies in the multi-ethnic meta-analysis of intracranial aneurysm in 8029 individuals
|
Study |
Nation |
Mean age (years)
|
Male
|
Sample size
|
|
Case |
Control |
Case |
Control |
Case |
Control |
|
Morgan et al. [28] (2006) |
GBR |
55 (24-80) |
56 (49-64) |
36 (40.0) |
2720 (100.0) |
91 |
2612 |
|
Slowik et al. [35] (2006) |
POL |
49.9±12.7 |
50.4±12.4 |
99 (42.9) |
99 (42.9) |
231 |
231 |
|
Fontanella et al. [9] (2007) |
ITA |
54.1±14.0 |
53.4±14.2 |
56 (32.8) |
70 (48.6) |
171 |
144 |
|
Fontanella et al. [8] (2008) |
ITA |
53.7±14.1 |
53.7±14.0 |
58 (32.4) |
50 (32.1) |
179 |
156 |
|
Sun et al. [37] (2008) |
CHN |
45.2±11.7 |
41.8±9.0 |
104 (43.3) |
116 (48.3) |
240 |
240 |
|
Fontanella et al. [10] (2010) |
ITA |
55.0±14.5 |
53.7±14.0 |
74 (34.4) |
50 (32.3) |
215 |
155 |
|
Zhang et al. [40] (2011) |
CHN |
36.0±4.2 |
33.0±4.5 |
103 (56.6) |
95 (52.2) |
182 |
182 |
|
Li et al. [22] (2012) |
CHN |
53.1±13.1 |
50.0±8.9 |
60 (36.6) |
108 (41.9) |
164 |
258 |
|
Liu et al. [24] (2012) |
CHN |
47.4±11.3 |
45.6±10.7 |
95 (43.2) |
103 (46.8) |
220 |
220 |
|
Bayri et al. [3] (2015) |
TUR |
- |
- |
- |
- |
120 |
120 |
|
Sathyan et al. [33] (2015) |
IND |
51.2±11.4 |
- |
123 (55.9) |
- |
226 |
244 |
|
Hendrix et al. [12] (2017) |
USA*
|
54.9±12.5 |
50.6±18.6 |
35 (23.5) |
18 (36.0) |
149 |
50 |
|
Borges et al. [4] (2018) |
BRA |
54.0±9.0 |
52.0±6.0 |
- |
38 (46.9) |
33 |
81 |
|
Xu et al. [39] (2021) |
CHN |
57.1±10.6 |
66.5±12.1 |
117 (30.5) |
117 (30.5) |
384 |
384 |
|
Hong et al. [13] (2022) |
KOR |
59.3±0.8 |
52.1±1.0 |
104 (41.6) |
142 (48.0) |
250 |
296 |
Table 2.
Summary of inflammation-related gene variants in intracranial aneurysm from each study enrolled in this meta-analysis
|
Study |
Gene |
SNP |
NE/E |
EAF |
Genotype*
|
OR (95% CI)†
|
|
Case |
Control |
|
Morgan et al. [28] (2006) |
IL-6 |
rs1800796 |
G/C |
0.051 |
79-8-4 |
2359/244/9 |
1.83 (1.08-3.10) |
|
Slowik et al. [35] (2006) |
IL-1β |
rs16944 |
C/T |
0.321 |
100/99/32 |
111/106/14 |
1.33 (1.01-1.76) |
|
Fontanella et al. [9] (2007) |
TNF-α |
rs1800629 |
G/A |
0.149 |
136/30/5 |
92/50/2 |
0.57 (0.37-0.89) |
|
Fontanella et al. [8] (2008) |
IL-6 |
rs1800796 |
G/C |
0.091 |
149/26/4 |
131/23/2 |
1.11 (0.65-1.88) |
|
Sun et al. [37] (2008) |
IL-6 |
rs1800796 |
G/C |
0.638 |
59/130/51 |
9/82/149 |
0.25 (0.19-0.33) |
|
Fontanella et al. [10] (2010) |
IL-1α |
rs1800587 |
C/T |
0.351 |
82/110/23 |
63/80/12 |
1.13 (0.83-1.53) |
|
Fontanella et al. [10] (2010) |
IL-1β |
rs16944 |
C/T |
0.362 |
94/88/33 |
64/68/23 |
0.96 (0.71-1.30) |
|
Zhang et al. [40] (2011) |
IL-6 |
rs1800796 |
G/C |
0.082 |
145/32/5 |
165/16/1 |
2.51 (1.41-4.45) |
|
Li et al. [22] (2012) |
IL-12B |
rs321222 |
A/C |
0.462 |
29/100/35 |
80/136/42 |
1.45 (1.10-1.91) |
|
Liu et al. [24] (2012) |
IL-6 |
rs1800796 |
G/C |
0.738 |
33/66/121 |
11/77/132 |
0.68 (0.50-0.92) |
|
Bayri et al. [3] (2015) |
IL-6 |
rs1800796 |
G/C |
0.144 |
94/24/2 |
83/33/4 |
0.64 (0.38-1.08) |
|
Sathyan et al. [33] (2015) |
TNF-α |
rs1800629 |
G/A |
0.100 |
185/41/0 |
192/51/1 |
0.82 (0.53-1.26) |
|
Sathyan et al. [33] (2015) |
IL-1α |
rs1800587 |
C/T |
0.302 |
109/83/27 |
118/108/17 |
0.91 (0.68-1.20) |
|
Sathyan et al. [33] (2015) |
IL-1β |
rs16944 |
C/T |
0.396 |
84/101/38 |
90/115/39 |
0.99 (0.76-1.29) |
|
Sathyan et al. [33] (2015) |
IL-6 |
rs1800796 |
G/C |
0.447 |
57/126/37 |
81/111/52 |
0.95 (0.73-1.23) |
|
Sathyan et al. [33] (2015) |
IL-12B |
rs321222 |
A/C |
0.384 |
88/96/36 |
83/115/31 |
0.98 (0.75-1.28) |
|
Hendrix et al. [12] (2017) |
HMGB1 |
rs2249825 |
C/G |
0.150 |
109/35/5 |
29/18/3 |
5.70 (1.80-17.98)‡
|
|
Borges et al. [4] (2018) |
TNF-α |
rs1800629 |
G/A |
0.325 |
21-3-9 |
47/15/19 |
0.96 (0.52-1.77) |
|
Xu et al. [39] (2021) |
TNF-α |
rs1800629 |
G/A |
0.059 |
337/46/1 |
342/41/1 |
1.14 (0.68-1.93) |
|
Xu et al. [39] (2021) |
TNF-α |
rs1799724 |
C/T |
0.124 |
293/86/5 |
298/77/9 |
0.97 (0.67-1.40) |
|
Xu et al. [39] (2021) |
TNF-α |
rs1799964 |
T/C |
0.178 |
272/102/10 |
252/112/20 |
0.71 (0.51-0.98) |
|
Xu et al. [39] (2021) |
IL-1α |
rs17561 |
C/A |
0.073 |
341/41/2 |
318/65/1 |
0.71 (0.44-1.15) |
|
Xu et al. [39] (2021) |
IL-1β |
rs1143627 |
G/A |
0.529 |
77/199/108 |
93/185/106 |
1.13 (0.88-1.45) |
|
Xu et al. [39] (2021) |
IL-1β |
rs16944 |
A/G |
0.544 |
71/196/117 |
88/187/109 |
1.18 (0.92-1.52) |
|
Xu et al. [39] (2021) |
IL-1β |
rs1143623 |
C/G |
0.401 |
134/185/65 |
144/179/61 |
0.98 (0.76-1.26) |
|
Xu et al. [39] (2021) |
IL-1β |
rs1143630 |
T/G |
0.831 |
12/108/264 |
13/102/269 |
1.10 (0.79-1.53) |
|
Xu et al. [39] (2021) |
IL-1β |
rs2853550 |
A/G |
0.907 |
0/58/326 |
5/75/304 |
1.31 (0.85-2.01) |
|
Xu et al. [39] (2021) |
IL-1β |
rs3136558 |
A/G |
0.342 |
160/176/48 |
171/172/41 |
0.91 (0.70-1.19) |
|
Xu et al. [39] (2021) |
IL-6 |
rs1800796 |
G/C |
0.773 |
17/137/230 |
18/141/225 |
1.02 (0.75-1.38) |
|
Xu et al. [39] (2021) |
IL-12B |
rs3181216 |
A/T |
0.282 |
207/152/25 |
191/155/38 |
0.74 (0.56-0.98) |
|
Xu et al. [39] (2021) |
IL-12B |
rs3212227 |
T/G |
0.473 |
105/191/88 |
107/195/82 |
1.04 (0.81-1.34) |
|
Xu et al. [39] (2021) |
IL-12B |
rs1003199 |
C/T |
0.378 |
131/208/45 |
148/190/46 |
1.13 (0.86-1.49) |
|
Xu et al. [39] (2021) |
IL-12B |
rs2195940 |
C/T |
0.050 |
354/28/2 |
341/41/2 |
0.59 (0.33-1.06) |
|
Hong et al. [13] (2022) |
TNF-α |
rs1800629 |
G/A |
0.060 |
220/30/0 |
261/34/1 |
1.15 (0.65-2.05) |
|
Hong et al. [13] (2022) |
TNF-α |
rs1799724 |
C/T |
0.181 |
158/89/3 |
201/87/8 |
1.24 (0.88-1.76) |
|
Hong et al. [13] (2022) |
TNF-α |
rs1799964 |
T/C |
0.212 |
143/96/11 |
192/95/9 |
1.33 (0.96-1.83) |
|
Hong et al. [13] (2022) |
IL-1α |
rs17561 |
C/A |
0.059 |
217/32/1 |
266/30/0 |
1.28 (0.73-2.23) |
|
Hong et al. [13] (2022) |
IL-1α |
rs1800587 |
C/T |
0.059 |
217/32/1 |
266/30/0 |
1.28 (0.73-2.23) |
|
Hong et al. [13] (2022) |
IL-1β |
rs2853550 |
G/A |
0.080 |
208/41/1 |
255/38/3 |
1.23 (0.77-1.97) |
|
Hong et al. [13] (2022) |
IL-1β |
rs3136558 |
A/G |
0.473 |
80/109/61 |
75/157/64 |
0.89 (0.69-1.15) |
|
Hong et al. [13] (2022) |
IL-1β |
rs1143630 |
G/T |
0.201 |
160/77/13 |
191/93/12 |
1.03 (0.75-1.41) |
|
Hong et al. [13] (2022) |
IL-1β |
rs1143627 |
A/G |
0.239 |
68/122/60 |
69/150/77 |
0.84 (0.65-1.08) |
|
Hong et al. [13] (2022) |
IL-1β |
rs1143623 |
C/G |
0.434 |
78/121/51 |
98/145/53 |
1.15 (0.89-1.48) |
|
Hong et al. [13] (2022) |
IL-1β |
rs16944 |
C/T |
0.498 |
60/122/68 |
79/148/69 |
1.21 (0.94-1.56) |
|
Hong et al. [13] (2022) |
IL-6 |
rs1800796 |
G/C |
0.239 |
148/82/20 |
174/105/17 |
0.90 (0.67-1.20) |
|
Hong et al. [13] (2022) |
IL-12B |
rs2195940 |
C/T |
0.052 |
228/20/2 |
263/33/0 |
0.72 (0.40-1.29) |
|
Hong et al. [13] (2022) |
IL-12B |
rs3181216 |
A/T |
0.271 |
117/113/20 |
170/109/17 |
1.34 (1.00-1.80) |
|
Hong et al. [13] (2022) |
IL-12B |
rs1003199 |
C/T |
0.318 |
107/112/31 |
147/125/24 |
1.31 (0.99-1.73) |
|
Hong et al. [13] (2022) |
IL-12B |
rs3212227 |
A/C |
0.486 |
70/106/74 |
84/147/65 |
0.90 (0.70-1.15) |
|
Hong et al. [13] (2022) |
HMGB1 |
rs2249825 |
C/G |
0.887 |
3/45/202 |
3/66/227 |
1.21 (0.78-1.89)‡
|
Table 3.
Results of meta-analysis of the inflammatory-related loci associated with intracranial aneurysm
|
Gene |
SNP |
NE |
E |
EAF |
Fixed effect*
|
Random effect*
|
I2*
|
Q value*
|
Sample
|
Study |
|
OR (95% CI) |
p-value |
OR (95% CI) |
p-value |
Case |
Control |
|
IL-1α |
rs17561 |
A |
C |
0.067 |
0.91 (0.63-1.31) |
0.6116 |
0.93 (0.53-1.66) |
0.8178 |
0.59 |
0.1178 |
634 |
680 |
2 [13,39] |
|
IL-1α |
rs1800587 |
C |
T |
0.219 |
1.04 (0.85-1.27) |
0.7105 |
1.04 (0.85-1.27) |
0.7105 |
0.00 |
0.4471 |
684 |
694 |
3 [10,13,33] |
|
IL-1β |
rs2853550 |
G |
A |
0.912 |
1.05 (0.77-1.45) |
0.7433 |
1.04 (0.65-1.66) |
0.8588 |
0.53 |
0.1458 |
634 |
680 |
2 [13,39] |
|
IL-1β |
rs3136558 |
G |
A |
0.396 |
0.90 (0.75-1.08) |
0.2534 |
0.90 (0.75-1.08) |
0.2534 |
0.00 |
0.9042 |
634 |
680 |
2 [13,39] |
|
IL-1β |
rs1143630 |
G |
T |
0.818 |
1.03 (0.82-1.29) |
0.7974 |
1.03 (0.82-1.29) |
0.7974 |
0.00 |
0.5936 |
634 |
680 |
2 [13,39] |
|
IL-1β |
rs1143627 |
A |
G |
0.625 |
1.16 (0.97-1.39) |
0.1039 |
1.16 (0.97-1.39) |
0.1039 |
0.00 |
0.7752 |
634 |
680 |
2 [13,39] |
|
IL-1β |
rs16944 |
C |
T |
0.443 |
1.13 (1.01-1.28) |
0.0390 |
1.13 (1.01-1.28) |
0.0390 |
0.00 |
0.4301 |
1303 |
1310 |
5 [10,13,33,35,39] |
|
IL-1β |
rs1143623 |
G |
C |
0.415 |
1.06 (0.88-1.27) |
0.5319 |
1.06 (0.88-1.27) |
0.5319 |
0.00 |
0.3969 |
634 |
680 |
2 [13,39] |
|
IL-12B |
rs3212227 |
A |
C |
0.475 |
1.04 (0.91-1.18) |
0.5587 |
1.05 (0.85-1.29) |
0.6576 |
0.59 |
0.0602 |
1018 |
1167 |
4 [13,22,33,39] |
|
IL-12B |
rs2195940 |
T |
C |
0.051 |
0.65 (0.43-0.98) |
0.0413 |
0.65 (0.43-0.98) |
0.0413 |
0.00 |
0.6318 |
634 |
680 |
2 [13,39] |
|
IL-12B |
rs3181216 |
T |
A |
0.277 |
0.98 (0.80-1.20) |
0.8439 |
0.99 (0.56-1.78) |
0.9834 |
0.88 |
0.0041 |
634 |
680 |
2 [13,39] |
|
IL-12B |
rs1003199 |
T |
C |
0.353 |
1.21 (1.00-1.48) |
0.0509 |
1.21 (1.00-1.48) |
0.0509 |
0.00 |
0.4619 |
634 |
680 |
2 [13,39] |
|
TNF-α |
rs1799964 |
C |
T |
0.192 |
0.98 (0.78-1.23) |
0.8655 |
0.97 (0.53-1.80) |
0.9285 |
0.86 |
0.0078 |
634 |
680 |
2 [13,39] |
|
TNF-α |
rs1799724 |
T |
C |
0.148 |
1.11 (0.86-1.43) |
0.4339 |
1.11 (0.86-1.43) |
0.4339 |
0.00 |
0.3375 |
634 |
680 |
2 [13,39] |
|
TNF-α |
rs1800629 |
G |
A |
0.618 |
0.78 (0.62-0.97) |
0.0260 |
0.78 (0.62-0.97) |
0.0260 |
0.00 |
0.5848 |
1064 |
1149 |
5 [4,9,13,33,39] |
|
IL-6 |
rs1800796 |
G |
C |
0.262 |
0.75 (0.67-0.85) |
1.81E-06 |
0.90 (0.59-1.37) |
0.6130 |
0.92 |
0.0000 |
1886 |
4454 |
9 [3,8,13,24,28,33,37,39,40] |
|
HMGB1†
|
rs2249825 |
C |
G |
0.690 |
1.47 (0.98-2.22) |
0.0645 |
2.39 (0.53-10.8) |
0.2578 |
0.84 |
0.0138 |
399 |
344 |
2 [12,13] |
Table 4.
Inflammation-related gene variants in the ethnicity-stratified trans-ethnic meta-analysis
|
Gene |
SNP |
NE |
E |
EAF |
Fixed effect*
|
Random effect*
|
I2*
|
Q value*
|
Sample
|
Study |
|
OR (95% CI) |
p-value |
OR (95% CI) |
p-value |
Case |
Control |
|
European-ancestry |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
IL-1β |
rs16944 |
C |
T |
0.339 |
1.15 (0.94-1.41) |
0.1863 |
1.14 (0.83-1.56) |
0.4310 |
0.59 |
0.1176 |
446 |
386 |
2 [10,35] |
|
IL-6 |
rs1800796 |
G |
C |
0.062 |
1.09 (0.80-1.47) |
0.5929 |
1.09 (0.60-1.98) |
0.7774 |
0.74 |
0.0211 |
390 |
2888 |
3 [3,8,28] |
|
East Asian |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
IL-1α |
rs17561 |
A |
C |
0.067 |
0.91 (0.63-1.31) |
0.6116 |
0.93 (0.53-1.66) |
0.8178 |
0.59 |
0.1178 |
634 |
680 |
2 [13,39] |
|
IL-1β |
rs2853550 |
G |
A |
0.912 |
1.05 (0.77-1.45) |
0.7433 |
1.04 (0.65-1.66) |
0.8588 |
0.53 |
0.1458 |
634 |
680 |
2 [13,39] |
|
IL-1β |
rs3136558 |
G |
A |
0.396 |
0.90 (0.75-1.08) |
0.2534 |
0.90 (0.75-1.08) |
0.2534 |
0.00 |
0.9042 |
634 |
680 |
2 [13,39] |
|
IL-1β |
rs1143630 |
G |
T |
0.818 |
1.03 (0.82-1.29) |
0.7974 |
1.03 (0.82-1.29) |
0.7974 |
0.00 |
0.5936 |
634 |
680 |
2 [13,39] |
|
IL-1β |
rs1143627 |
A |
G |
0.625 |
1.16 (0.97-1.39) |
0.1039 |
1.16 (0.97-1.39) |
0.1039 |
0.00 |
0.7752 |
634 |
680 |
2 [13,39] |
|
IL-1β |
rs16944 |
C |
T |
0.525 |
1.19 (1.00-1.43) |
0.0492 |
1.19 (1.00-1.43) |
0.0492 |
0.00 |
0.8896 |
634 |
680 |
2 [13,39] |
|
IL-1β |
rs1143623 |
G |
C |
0.415 |
1.06 (0.88-1.27) |
0.5319 |
1.06 (0.88-1.27) |
0.5319 |
0.00 |
0.3969 |
634 |
680 |
2 [13,39] |
|
IL-12B |
rs3212227 |
A |
C |
0.498 |
1.06 (0.91-1.23) |
0.4520 |
1.07 (0.81-1.42) |
0.6263 |
0.72 |
0.0280 |
1018 |
1167 |
3 [13,16,22] |
|
IL-12B |
rs2195940 |
T |
C |
0.051 |
0.65 (0.43-0.98) |
0.0413 |
0.65 (0.43-0.98) |
0.0413 |
0.00 |
0.6318 |
634 |
680 |
2 [13,39] |
|
IL-12B |
rs3181216 |
T |
A |
0.277 |
0.98 (0.80-1.20) |
0.8439 |
0.99 (0.56-1.78) |
0.9834 |
0.88 |
0.0041 |
634 |
680 |
2 [13,39] |
|
IL-12B |
rs1003199 |
T |
C |
0.353 |
1.21 (1.00-1.48) |
0.0509 |
1.21 (1.00-1.48 |
0.0509 |
0.00 |
0.4619 |
634 |
680 |
2 [13,39] |
|
TNF-α |
rs1799964 |
C |
T |
0.192 |
0.98 (0.78-1.23) |
0.8655 |
0.97 (0.53-1.80) |
0.9285 |
0.86 |
0.0078 |
634 |
680 |
2 [13,39] |
|
TNF-α |
rs1799724 |
T |
C |
0.148 |
1.11 (0.86-1.43) |
0.4339 |
1.11 (0.86-1.43) |
0.4339 |
0.00 |
0.3375 |
634 |
680 |
2 [13,39] |
|
TNF-α |
rs1800629 |
A |
G |
0.059 |
1.15 (0.78-1.68) |
0.4855 |
1.15 (0.78-1.68) |
0.4855 |
0.00 |
0.9753 |
634 |
680 |
2 [13,39] |
|
IL-6 |
rs1800796 |
G |
C |
0.481 |
0.65 (0.56-0.75) |
3.24E-09 |
0.80 (0.42-1.54) |
0.5062 |
0.95 |
2.22E-16 |
1276 |
1322 |
5 [13,24,37,39,40] |
|
East + South Asian |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
IL-1α |
rs1800587 |
C |
T |
0.170 |
0.98 (0.76-1.27) |
0.8678 |
0.99 (0.74-1.32) |
0.9365 |
0.10 |
0.2905 |
469 |
539 |
2 [13,33] |
|
IL-1β |
rs16944 |
C |
T |
0.491 |
1.13 (0.97-1.31) |
0.1113 |
1.13 (0.97-1.31) |
0.1113 |
0.00 |
0.5070 |
857 |
924 |
3 [13,33,39] |
|
IL-12B |
rs3212227 |
A |
C |
0.475 |
1.04 (0.91-1.18) |
0.5587 |
1.05 (0.85-1.29) |
0.6576 |
0.59 |
0.0602 |
1018 |
1167 |
4 [13,22,33,39] |
|
TNF-α |
rs1800629 |
A |
G |
0.281 |
1.18 (0.88-1.57) |
0.2660 |
1.18 (0.88-1.57) |
0.2660 |
0.00 |
0.9779 |
860 |
924 |
3 [13,33,39] |
|
IL-6 |
rs1800796 |
G |
C |
0.476 |
0.71 (0.62-0.80) |
7.28E-08 |
0.82 (0.48-1.40) |
0.4668 |
0.94 |
0.0000 |
1496 |
1566 |
6 [13,24,33,37,39,40] |
References
1. Ahn JH, Kim HC, Rhim JK, Park JJ, Sigmund D, Park MC, et al : Multiview convolutional neural networks in rupture risk assessment of small, unruptured intracranial aneurysms. J Pers Med 11 : 239, 2021    3. Bayri Y, Taskin E, Ulus A, Bayrakli F, Altun A, Bagci H : Lack of association between interleukin 6 gene promoter polymorphisms and aneurysmal subarachnoid hemorrhage in Turkish population. J Neurol Sci 32 : 288-292, 2015
4. Borges FS, Freitas RS, Morais RM, Funghetto SS, Nóbrega OT, Ferreira LB, et al : TNFA gene in Brazilian patients with hemorrhagic stroke or cerebral aneurysm. J Bras Patol Med Lab 54 : 164-169, 2018
6. Chen Z, Song S, Zhu J, Lai X : Regulatory mechanism of MiR-21 in formation and rupture of intracranial aneurysm through JNK signaling pathway-mediated inflammatory response. Int J Clin Exp Pathol 13 : 1834-1841, 2020   7. Connolly ES Jr, Choudhri TF, Mack WJ, Mocco J, Spinks TJ, Slosberg J, et al : Influence of smoking, hypertension, and sex on the phenotypic expression of familial intracranial aneurysms in siblings. Neurosurgery 48 : 64-68; discussion 68-69, 2001   8. Fontanella M, Rainero I, Gallone S, Rubino E, Fenoglio P, Valfrè W, et al : Interleukin 6 gene polymorphisms are not associated with aneurysmal subarachnoid haemorrhage in an Italian population. J Neurol Neurosurg Psychiatry 79 : 471-473, 2008   9. Fontanella M, Rainero I, Gallone S, Rubino E, Fenoglio P, Valfrè W, et al : Tumor necrosis factor-alpha gene and cerebral aneurysms. Neurosurgery 60 : 668-672; discussion 672-673, 2007  10. Fontanella M, Rainero I, Gallone S, Rubino E, Fornaro R, Fenoglio P, et al : Interleukin-1 cluster gene polymorphisms and aneurysmal subarachnoid hemorrhage. Neurosurgery 66 : 1058-1062; discussion 1062-1063, 2010   11. Han SW, Kim BJ, Kim TY, Lim SH, Youn DH, Hong EP, et al : Association of haptoglobin phenotype with neurological and cognitive outcomes in patients with subarachnoid hemorrhage. Front Aging Neurosci 14 : 819628, 2022    12. Hendrix P, Foreman PM, Harrigan MR, Fisher WS Rd, Vyas NA, Lipsky RH, et al : Impact of high-mobility group box 1 polymorphism on delayed cerebral ischemia after aneurysmal subarachnoid hemorrhage. World Neurosurg 101 : 325-330, 2017   13. Hong EP, Kim BJ, Youn DH, Lee JJ, Jeon HJ, Choi HJ, et al : Updated genome-wide association study of intracranial aneurysms by genotype correction and imputation in Koreans. World Neurosurg 166 : e109-e117, 2022   14. Hong EP, Youn DH, Kim BJ, Lee JJ, Na D, Ahn JH, et al : Genome-wide polygenic risk impact on intracranial aneurysms and acute ischemic stroke. PLoS One 17 : e02655812022    15. Hong EP, Youn DH, Kim BJ, Lee JJ, Nam S, Yoo H, et al : Novel genomewide interactions mediated via BOLL and EDNRA polymorphisms in intracranial aneurysm. J Korean Neurosurg Soc 2022, [Epub ahead of print]  17. Jayaraman T, Berenstein V, Li X, Mayer J, Silane M, Shin YS, et al : Tumor necrosis factor alpha is a key modulator of inflammation in cerebral aneurysms. Neurosurgery 57 : 558-564; discussion 558-564, 2005  18. Johnston WF, Salmon M, Pope NH, Meher A, Su G, Stone ML, et al : Inhibition of interleukin-1β decreases aneurysm formation and progression in a novel model of thoracic aortic aneurysms. Circulation 130( 11 Suppl 1):S51-S59, 2014    19. Kao HW, Lee KW, Kuo CL, Huang CS, Tseng WM, Liu CS, et al : Interleukin-6 as a prognostic biomarker in ruptured intracranial aneurysms. PLoS One 10 : e01321152015    20. Kataoka H : Molecular mechanisms of the formation and progression of intracranial aneurysms. Neurol Med Chir (Tokyo) 55 : 214-229, 2015    22. Li LJ, Pan XM, Sima X, Li ZH, Zhang LS, Sun H, et al : Interactions of interleukin-12A and interleukin-12B polymorphisms on the risk of intracranial aneurysm. Mol Biol Rep 39 : 11217-11223, 2012    24. Liu Y, Sun J, Wu C, Cao X, He M, You C : The interleukin-6-572G/C gene polymorphism and the risk of intracranial aneurysms in a Chinese population. Genet Test Mol Biomarkers 16 : 822-826, 2012   26. McColgan P, Thant KZ, Sharma P : The genetics of sporadic ruptured and unruptured intracranial aneurysms: a genetic meta-analysis of 8 genes and 13 polymorphisms in approximately 20,000 individuals. J Neurosurg 112 : 714-721, 2010   27. Monsour M, Croci DM, Grüter BE, Taussky P, Marbacher S, Agazzi S : Cerebral aneurysm and interleukin-6: a key player in aneurysm generation and rupture or just one of the multiple factors? Transl Stroke Res 2022, [Epub ahead of print]   28. Morgan L, Cooper J, Montgomery H, Kitchen N, Humphries SE : The interleukin-6 gene -174G>C and -572G>C promoter polymorphisms are related to cerebral aneurysms. J Neurol Neurosurg Psychiatry 77 : 915-917, 2006    29. Nemetz A, Nosti-Escanilla MP, Molnár T, Köpe A, Kovács A, Fehér J, et al : IL1B gene polymorphisms influence the course and severity of inflammatory bowel disease. Immunogenetics 49 : 527-531, 1999    30. Nin JW, Ferreira I, Schalkwijk CG, Jorsal A, Prins MH, Parving HH, et al : Higher plasma high-mobility group box 1 levels are associated with incident cardiovascular disease and all-cause mortality in type 1 diabetes: a 12 year follow-up study. Diabetologia 55 : 2489-2493, 2012    32. Pola R, Gaetani E, Flex A, Aloi F, Papaleo P, Gerardino L, et al : -174 G/C interleukin-6 gene polymorphism and increased risk of multi-infarct dementia: a case-control study. Exp Gerontol 37 : 949-955, 2002   34. Sawyer DM, Pace LA, Pascale CL, Kutchin AC, O’Neill BE, Starke RM, et al : Lymphocytes influence intracranial aneurysm formation and rupture: role of extracellular matrix remodeling and phenotypic modulation of vascular smooth muscle cells. J Neuroinflammation 13 : 185, 2016    35. Slowik A, Borratynska A, Turaj W, Pera J, Dziedzic T, Wloch D, et al : Interleukin 1beta-511 C/T polymorphism and risk of aneurysmal subarachnoid haemorrhage. J Neurol Neurosurg Psychiatry 77 : 279-280, 2006   36. Starke RM, Chalouhi N, Jabbour PM, Tjoumakaris SI, Gonzalez LF, Rosenwasser RH, et al : Critical role of TNF-α in cerebral aneurysm formation and progression to rupture. J Neuroinflammation 11 : 77, 2014    37. Sun H, Zhang D, Zhao J : The interleukin-6 gene -572G>C promoter polymorphism is related to intracranial aneurysms in Chinese Han nationality. Neurosci Lett 440 : 1-3, 2008   38. Tsai YC, Liu CJ, Huang HC, Lin JH, Chen PY, Su YK, et al : A meta-analysis of dynamic prevalence of cognitive deficits in the acute, subacute, and chronic phases after traumatic brain injury. J Neurosci Nurs 53 : 63-68, 2021   40. Zhang G, Tu Y, Feng W, Huang L, Li M, Qi S : Association of interleukin-6-572G/C gene polymorphisms in the Cantonese population with intracranial aneurysms. J Neurol Sci 306 : 94-97, 2011  
|
|


















